The Multimodal Brain Tumor Image Segmentation Benchmark (BRATS)
- PMID: 25494501
- PMCID: PMC4833122
- DOI: 10.1109/TMI.2014.2377694
The Multimodal Brain Tumor Image Segmentation Benchmark (BRATS)
Abstract
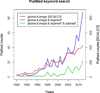
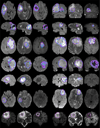
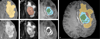
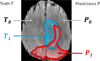
In this paper we report the set-up and results of the Multimodal Brain Tumor Image Segmentation Benchmark (BRATS) organized in conjunction with the MICCAI 2012 and 2013 conferences. Twenty state-of-the-art tumor segmentation algorithms were applied to a set of 65 multi-contrast MR scans of low- and high-grade glioma patients-manually annotated by up to four raters-and to 65 comparable scans generated using tumor image simulation software. Quantitative evaluations revealed considerable disagreement between the human raters in segmenting various tumor sub-regions (Dice scores in the range 74%-85%), illustrating the difficulty of this task. We found that different algorithms worked best for different sub-regions (reaching performance comparable to human inter-rater variability), but that no single algorithm ranked in the top for all sub-regions simultaneously. Fusing several good algorithms using a hierarchical majority vote yielded segmentations that consistently ranked above all individual algorithms, indicating remaining opportunities for further methodological improvements. The BRATS image data and manual annotations continue to be publicly available through an online evaluation system as an ongoing benchmarking resource.
Figures







References
-
- Holland EC. Progenitor cells and glioma formation. Curr. Opin. Neurol. 2001;14:683–688. - PubMed
-
- Ohgaki H, Kleihues P. Population-based studies on incidence, survival rates, and genetic alterations in astrocytic and oligodendroglial gliomas. J Neuropathol. Exp. Neurol. 2005 Jun;64(6):479–489. - PubMed
-
- Eisenhauer E, et al. New response evaluation criteria in solid tumours: Revised RECIST guideline (version 1.1) Eur. J. Cancer. 2009;45(2):228–247. - PubMed
-
- Wen PY, et al. Updated response assessment criteria for high-grade gliomas: Response assessment in neuro-oncology working group. J Clin. Oncol. 2010;28:1963–1972. - PubMed
Publication types
MeSH terms
Grants and funding
- P41-RR14075/RR/NCRR NIH HHS/United States
- P41-RR13218/RR/NCRR NIH HHS/United States
- P41-EB-015902/EB/NIBIB NIH HHS/United States
- U54 EB005149/EB/NIBIB NIH HHS/United States
- P41 RR013218/RR/NCRR NIH HHS/United States
- R01 EB013565/EB/NIBIB NIH HHS/United States
- U01 CA154601/CA/NCI NIH HHS/United States
- R01EB013565/EB/NIBIB NIH HHS/United States
- R15CA115464/CA/NCI NIH HHS/United States
- NIHR/CS/009/011/DH_/Department of Health/United Kingdom
- P41 EB015902/EB/NIBIB NIH HHS/United States
- U54-EB005149/EB/NIBIB NIH HHS/United States
- P41 RR014075/RR/NCRR NIH HHS/United States
- R15 CA115464/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

