ARHGEF3 controls HDACi-induced differentiation via RhoA-dependent pathways in acute myeloid leukemias
- PMID: 25494542
- PMCID: PMC4622697
- DOI: 10.4161/15592294.2014.988035
ARHGEF3 controls HDACi-induced differentiation via RhoA-dependent pathways in acute myeloid leukemias
Abstract
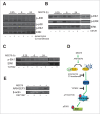
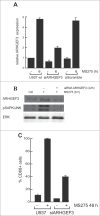
Altered expression and activity of histone deacetylases (HDACs) have been correlated with tumorigenesis. Inhibitors of HDACs (HDACi) induce acetylation of histone and non-histone proteins affecting gene expression, cell cycle progression, cell migration, terminal differentiation and cell death. Here, we analyzed the regulation of ARHGEF3, a RhoA-specific guanine nucleotide exchange factor, by the HDACi MS275 (entinostat). MS275 is a well-known benzamide-based HDACi, which induces differentiation of the monoblastic-like human histiocytic lymphoma cell line U937 to monocytes/macrophages. Incubation of U937 cells with MS275 resulted in an up regulation of ARHGEF3, followed by a significant enhancement of the marker of macrophage differentiation CD68. ARHGEF3 protein is primarily nuclear, but MS275 treatment rapidly induced its translocation into the cytoplasm. ARHGEF3 cytoplasmic localization is associated with activation of the RhoA/Rho-associated Kinase (ROCK) pathway. In addition to cytoskeletal rearrangements orchestrated by RhoA, we showed that ARHGEF3/RhoA-dependent signals involve activation of SAPK/JNK and then Elk1 transcription factor. Importantly, MS275-induced CD68 expression was blocked by exposure of U937 cells to exoenzyme C3 transferase and Y27632, inhibitors of Rho and ROCK respectively. Moreover, ARHGEF3 silencing prevented RhoA activation leading to a reduction in SAPK/JNK phosphorylation, Elk1 activation and CD68 expression, suggesting a crucial role for ARHGEF3 in myeloid differentiation. Taken together, our results demonstrate that ARHGEF3 modulates acute myeloid leukemia differentiation through activation of RhoA and pathways directly controlled by small GTPase family proteins. The finding that GEF protein modulation by HDAC inhibition impacts on cell differentiation may be important for understanding the antitumor mechanism(s) by which HDACi treatment stimulates differentiation in cancer.
Keywords: HDAC inhibitors; acute myeloid leukemia; cancer; differentiation; signal transduction.
Figures
References
-
- Conte M, Altucci L. Molecular pathways: the complexity of the epigenome in cancer and recent clinical advances. Clin Cancer Res 2012; 18:5526-34; PMID:22904103; http://dx.doi.org/10.1158/1078-0432.CCR-12-2037 - DOI - PubMed
-
- Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 2009; 325:834-40; PMID:19608861; http://dx.doi.org/10.1126/science.1175371 - DOI - PubMed
-
- Di Costanzo A, Del Gaudio N, Migliaccio A, Altucci L. Epigenetic drugs against cancer: an evolving landscape. Arch Toxicol 2014; 88:1651-68; PMID:25085708; http://dx.doi.org/10.1007/s00204-014-1315-6 - DOI - PubMed
-
- Mai A, Altucci L. Epi-drugs to fight cancer: from chemistry to cancer treatment, the road ahead. Int J Biochem Cell Biol 2009; 41:199-213; PMID:18790076; http://dx.doi.org/10.1016/j.biocel.2008.08.020 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous
