An improved genome of the model marine alga Ostreococcus tauri unfolds by assessing Illumina de novo assemblies
- PMID: 25494611
- PMCID: PMC4378021
- DOI: 10.1186/1471-2164-15-1103
An improved genome of the model marine alga Ostreococcus tauri unfolds by assessing Illumina de novo assemblies
Abstract
Background: Cost effective next generation sequencing technologies now enable the production of genomic datasets for many novel planktonic eukaryotes, representing an understudied reservoir of genetic diversity. O. tauri is the smallest free-living photosynthetic eukaryote known to date, a coccoid green alga that was first isolated in 1995 in a lagoon by the Mediterranean sea. Its simple features, ease of culture and the sequencing of its 13 Mb haploid nuclear genome have promoted this microalga as a new model organism for cell biology. Here, we investigated the quality of genome assemblies of Illumina GAIIx 75 bp paired-end reads from Ostreococcus tauri, thereby also improving the existing assembly and showing the genome to be stably maintained in culture.
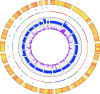
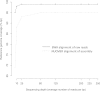
Results: The 3 assemblers used, ABySS, CLCBio and Velvet, produced 95% complete genomes in 1402 to 2080 scaffolds with a very low rate of misassembly. Reciprocally, these assemblies improved the original genome assembly by filling in 930 gaps. Combined with additional analysis of raw reads and PCR sequencing effort, 1194 gaps have been solved in total adding up to 460 kb of sequence. Mapping of RNAseq Illumina data on this updated genome led to a twofold reduction in the proportion of multi-exon protein coding genes, representing 19% of the total 7699 protein coding genes. The comparison of the DNA extracted in 2001 and 2009 revealed the fixation of 8 single nucleotide substitutions and 2 deletions during the approximately 6000 generations in the lab. The deletions either knocked out or truncated two predicted transmembrane proteins, including a glutamate-receptor like gene.
Conclusion: High coverage (>80 fold) paired-end Illumina sequencing enables a high quality 95% complete genome assembly of a compact ~13 Mb haploid eukaryote. This genome sequence has remained stable for 6000 generations of lab culture.
Figures

Similar articles
-
The genome of flax (Linum usitatissimum) assembled de novo from short shotgun sequence reads.Plant J. 2012 Nov;72(3):461-73. doi: 10.1111/j.1365-313X.2012.05093.x. Epub 2012 Aug 14. Plant J. 2012. PMID: 22757964
-
Screening the Sargasso Sea metagenome for data to investigate genome evolution in Ostreococcus (Prasinophyceae, Chlorophyta).Gene. 2007 Dec 30;406(1-2):184-90. doi: 10.1016/j.gene.2007.09.015. Epub 2007 Oct 3. Gene. 2007. PMID: 17961934
-
Ostreococcus tauri is a new model green alga for studying iron metabolism in eukaryotic phytoplankton.BMC Genomics. 2016 May 3;17:319. doi: 10.1186/s12864-016-2666-6. BMC Genomics. 2016. PMID: 27142620 Free PMC article.
-
Ostreococcus tauri: seeing through the genes to the genome.Trends Genet. 2007 Apr;23(4):151-4. doi: 10.1016/j.tig.2007.02.008. Epub 2007 Feb 28. Trends Genet. 2007. PMID: 17331615 Review.
-
The tiny giant of the sea, Ostreococcus's unique adaptations.Plant Physiol Biochem. 2024 Jun;211:108661. doi: 10.1016/j.plaphy.2024.108661. Epub 2024 May 1. Plant Physiol Biochem. 2024. PMID: 38735153 Review.
Cited by
-
Jabba: hybrid error correction for long sequencing reads.Algorithms Mol Biol. 2016 May 3;11:10. doi: 10.1186/s13015-016-0075-7. eCollection 2016. Algorithms Mol Biol. 2016. PMID: 27148393 Free PMC article.
-
Fossil-calibrated molecular clock data enable reconstruction of steps leading to differentiated multicellularity and anisogamy in the Volvocine algae.BMC Biol. 2024 Apr 10;22(1):79. doi: 10.1186/s12915-024-01878-1. BMC Biol. 2024. PMID: 38600528 Free PMC article.
-
Deep-coverage spatiotemporal proteome of the picoeukaryote Ostreococcus tauri reveals differential effects of environmental and endogenous 24-hour rhythms.Commun Biol. 2021 Sep 30;4(1):1147. doi: 10.1038/s42003-021-02680-3. Commun Biol. 2021. PMID: 34593975 Free PMC article.
-
Population genomics of picophytoplankton unveils novel chromosome hypervariability.Sci Adv. 2017 Jul 5;3(7):e1700239. doi: 10.1126/sciadv.1700239. eCollection 2017 Jul. Sci Adv. 2017. PMID: 28695208 Free PMC article.
-
Biochemical Characterization and Crystal Structure of a Novel NAD+-Dependent Isocitrate Dehydrogenase from Phaeodactylum tricornutum.Int J Mol Sci. 2020 Aug 18;21(16):5915. doi: 10.3390/ijms21165915. Int J Mol Sci. 2020. PMID: 32824636 Free PMC article.
References
-
- Karsenti E, Acinas SG, Bork P, Bowler C, De Vargas C, Raes J, Sullivan M, Arendt D, Benzoni F, Claverie J-M, Follows M, Gorsky G, Hingamp P, Iudicone D, Jaillon O, Kandels-Lewis S, Krzic U, Not F, Ogata H, Pesant S, Reynaud EG, Sardet C, Sieracki ME, Speich S, Velayoudon D, Weissenbach J, Wincker P, Consortium Tara Oceans A holistic approach to marine eco-systems biology. PLoS Biol. 2011;9:e1001177. doi: 10.1371/journal.pbio.1001177. - DOI - PMC - PubMed
-
- Keeling PJ, Burki F, Wilcox HM, Allam B, Allen EE, Amaral-Zettler LA, Armbrust EV, Archibald JM, Bharti AK, Bell CJ, Beszteri B, Bidle KD, Cameron CT, Campbell L, Caron DA, Cattolico RA, Collier JL, Coyne K, Davy SK, Deschamps P, Dyhrman ST, Edvardsen B, Gates RD, Gobler CJ, Greenwood SJ, Guida SM, Jacobi JL, Jakobsen KS, James ER, Jenkins B, et al. The marine microbial eukaryote transcriptome sequencing project (MMETSP): illuminating the functional diversity of eukaryotic life in the oceans through transcriptome sequencing. PLoS Biol. 2014;12:e1001889. doi: 10.1371/journal.pbio.1001889. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

