Nucleosome structure and function
- PMID: 25495456
- PMCID: PMC4378457
- DOI: 10.1021/cr500373h
Nucleosome structure and function
Figures

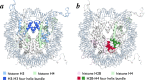

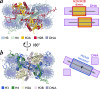



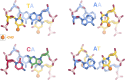


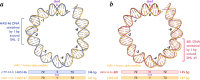


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

