Production of a Chaetomium globosum enolase monoclonal antibody
- PMID: 25495488
- PMCID: PMC4278173
- DOI: 10.1089/mab.2014.0042
Production of a Chaetomium globosum enolase monoclonal antibody
Abstract
Chaetomium globosum is a hydrophilic fungal species and a contaminant of water-damaged building materials in North America. Methods to detect Chaetomium species include subjective identification of ascospores, viable culture, or molecular-based detection methods. In this study, we describe the production and initial characterization of a monoclonal antibody (MAb) for C. globosum enolase. MAb 1C7, a murine IgG1 isotype MAb, was produced and reacted with recombinant C. globosum enolase (rCgEno) in an enzyme-linked immunosorbent assay and with a putative C. globosum enolase in a Western blot. Epitope mapping showed MAb 1C7 specific reactivity to an enolase decapeptide, LTYEELANLY, that is highly conserved within the fungal class Sordariomycetes. Cross-reactivity studies showed MAb 1C7 reactivity to C. atrobrunneum but not C. indicum. MAb 1C7 did not react with enolase from Aspergillus fumigatus, which is divergent in only two amino acids within this epitope. The results of this study suggest potential utility of MAb 1C7 in Western blot applications for the detection of Chaetomium and other Sordariomycetes species.
Figures


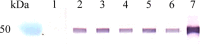


Similar articles
-
Characterization of an antigenic chitosanase from the cellulolytic fungus Chaetomium globosum.Med Mycol. 2013 Apr;51(3):290-9. doi: 10.3109/13693786.2012.715246. Epub 2012 Sep 18. Med Mycol. 2013. PMID: 22985087
-
Identification of an epitope of alpha-enolase (a candidate plasminogen receptor) by phage display.Thromb Haemost. 1997 Sep;78(3):1097-103. Thromb Haemost. 1997. PMID: 9308760
-
Quantification of C. globosum spores in house dust samples.Ann Agric Environ Med. 2014;21(3):525-30. doi: 10.5604/12321966.1120595. Ann Agric Environ Med. 2014. PMID: 25292122
-
Production and characterization of murine monoclonal antibody against synthetic peptide of CD34.Hum Antibodies. 2013;22(1-2):1-8. doi: 10.3233/HAB-130265. Hum Antibodies. 2013. PMID: 24284303 Review.
-
Invasive pulmonary mycosis due to Chaetomium globosum with false-positive galactomannan test: a case report and literature review.Mycoses. 2016 Mar;59(3):186-93. doi: 10.1111/myc.12446. Epub 2015 Dec 22. Mycoses. 2016. PMID: 26691935 Review.
Cited by
-
The Sordariomycetes: an expanding resource with Big Data for mining in evolutionary genomics and transcriptomics.Front Fungal Biol. 2023 Jun 30;4:1214537. doi: 10.3389/ffunb.2023.1214537. eCollection 2023. Front Fungal Biol. 2023. PMID: 37746130 Free PMC article. Review.
-
Integrative Activity of Mating Loci, Environmentally Responsive Genes, and Secondary Metabolism Pathways during Sexual Development of Chaetomium globosum.mBio. 2019 Dec 10;10(6):e02119-19. doi: 10.1128/mBio.02119-19. mBio. 2019. PMID: 31822585 Free PMC article.
References
-
- Horner WE, Barnes C, Codina R, and Levetin E: Guide for interpreting reports from inspections/investigations of indoor mold. J Allergy Clin Immunol 2008;121:592–597;e597 - PubMed
-
- Brasel TL, Campbell AW, Demers RE, Ferguson BS, Fink J, Vojdani A, Wilson SC, and Straus DC: Detection of trichothecene mycotoxins in sera from individuals exposed to Stachybotrys chartarum in indoor environments. Arch Environ Health 2004;59:317–323 - PubMed
-
- Fogle MR, Douglas DR, Jumper CA, and Straus DC: Growth and mycotoxin production by Chaetomium globosum. Mycopathologia 2007;164:49–56 - PubMed
-
- Provost NB, Shi C, She YM, Cyr TD, and Miller JD: Characterization of an antigenic chitosanase from the cellulolytic fungus Chaetomium globosum. Med Mycol 2013;51:290–299 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
