Autophagy fosters myofibroblast differentiation through MTORC2 activation and downstream upregulation of CTGF
- PMID: 25495560
- PMCID: PMC4502773
- DOI: 10.4161/15548627.2014.981786
Autophagy fosters myofibroblast differentiation through MTORC2 activation and downstream upregulation of CTGF
Abstract
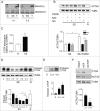
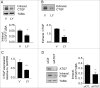
Recent evidence suggests that autophagy may favor fibrosis through enhanced differentiation of fibroblasts in myofibroblasts. Here, we sought to characterize the mediators and signaling pathways implicated in autophagy-induced myofibroblast differentiation. Fibroblasts, serum starved for up to 4 d, showed increased LC3-II/-I ratios and decreased SQSTM1/p62 levels. Autophagy was associated with acquisition of markers of myofibroblast differentiation including increased protein levels of ACTA2/αSMA (actin, α 2, smooth muscle, aorta), enhanced gene and protein levels of COL1A1 (collagen, type I, α 1) and COL3A1, and the formation of stress fibers. Inhibiting autophagy with 3 different class I phosphoinositide 3-kinase and class III phosphatidylinositol 3-kinase (PtdIns3K) inhibitors or through ATG7 silencing prevented myofibroblast differentiation. Autophagic fibroblasts showed increased expression and secretion of CTGF (connective tissue growth factor), and CTGF silencing prevented myofibroblast differentiation. Phosphorylation of the MTORC1 target RPS6KB1/p70S6K kinase was abolished in starved fibroblasts. Phosphorylation of AKT at Ser473, a MTORC2 target, was reduced after initiation of starvation but was followed by spontaneous rephosphorylation after 2 d of starvation, suggesting the reactivation of MTORC2 with sustained autophagy. Inhibiting MTORC2 activation with long-term exposure to rapamycin or by silencing RICTOR, a central component of the MTORC2 complex abolished AKT rephosphorylation. Both RICTOR silencing and rapamycin treatment prevented CTGF and ACTA2 upregulation, demonstrating the central role of MTORC2 activation in CTGF induction and myofibroblast differentiation. Finally, inhibition of autophagy with PtdIns3K inhibitors or ATG7 silencing blocked AKT rephosphorylation. Collectively, these results identify autophagy as a novel activator of MTORC2 signaling leading to CTGF induction and myofibroblast differentiation.
Keywords: 3-MA, 3-methyladenine; ACTA2, actin, α 2, smooth muscle, aorta; AKT; ATG7; Ctl, control; DAPI, 4′, 6-diamidino-2-phenylindole; ECM, extracellular matrix; FBS, fetal bovine serum; GF, growth factor; LC3B, MAP1LC3B (microtubule-associated protein 1 light chain 3 β); LY, LY294002; MTORC2; N, normal growth medium; R, rapamycin; SS, serum-free (starvation) medium; T, TGFB1 (transforming growth factor, β 1); TUBA, tubulin, α; V, vehicle; W, wortmannin; WB, western blotting; autophagy; connective tissue growth factor (CTGF); differentiation; fibroblast; fibrosis; iso, isotype control; myofibroblast; rapamycin.
Figures






References
-
- Hinz B. Formation and function of the myofibroblast during tissue repair. J Invest Dermatol 2007; 127:526-37; PMID:17299435; http://dx.doi.org/10.1038/sj.jid.5700613 - DOI - PubMed
-
- Wynn TA. Common and unique mechanisms regulate fibrosis in various fibroproliferative diseases. J Clin Invest 2007; 117:524-9; PMID:17332879; http://dx.doi.org/10.1172/JCI31487 - DOI - PMC - PubMed
-
- Tomasek JJ, Gabbiani G, Hinz B, Chaponnier C, Brown RA. Myofibroblasts and mechano-regulation of connective tissue remodelling. Nat Rev Mol Cell Bio 2002; 3:349-63; http://dx.doi.org/10.1038/nrm809 - DOI - PubMed
-
- Hinz B, Gabbiani G. Fibrosis: recent advances in myofibroblast biology and new therapeutic perspectives. F1000 Biol Rep 2010; 2:78; PMID:21170369; http://dx.doi.org/10.3410/B2-78 - DOI - PMC - PubMed
-
- Desmouliere A, Darby IA, Gabbiani G. Normal and pathologic soft tissue remodeling: role of the myofibroblast, with special emphasis on liver and kidney fibrosis. Lab Invest 2003; 83:1689-707; PMID:14691287; http://dx.doi.org/10.1097/01.LAB.0000101911.53973.90 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
