Methodology for the inference of gene function from phenotype data
- PMID: 25495798
- PMCID: PMC4302099
- DOI: 10.1186/s12859-014-0405-z
Methodology for the inference of gene function from phenotype data
Abstract
Background: Biomedical ontologies are increasingly instrumental in the advancement of biological research primarily through their use to efficiently consolidate large amounts of data into structured, accessible sets. However, ontology development and usage can be hampered by the segregation of knowledge by domain that occurs due to independent development and use of the ontologies. The ability to infer data associated with one ontology to data associated with another ontology would prove useful in expanding information content and scope. We here focus on relating two ontologies: the Gene Ontology (GO), which encodes canonical gene function, and the Mammalian Phenotype Ontology (MP), which describes non-canonical phenotypes, using statistical methods to suggest GO functional annotations from existing MP phenotype annotations. This work is in contrast to previous studies that have focused on inferring gene function from phenotype primarily through lexical or semantic similarity measures.
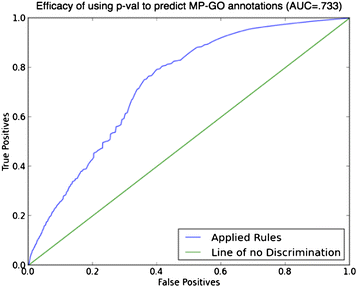
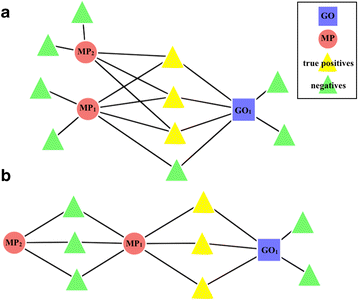
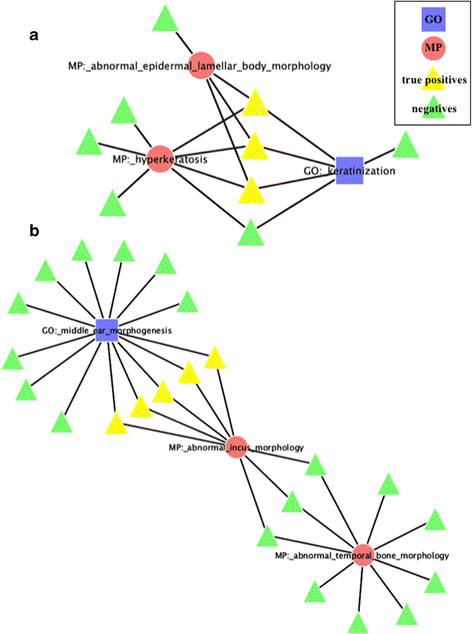
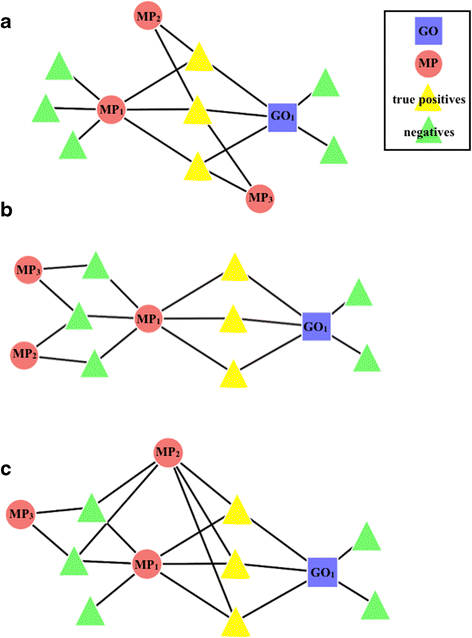
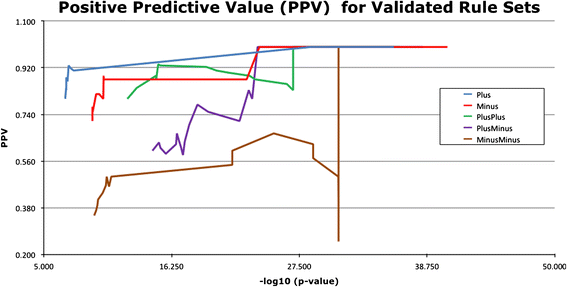
Results: We have designed and tested a set of algorithms that represents a novel methodology to define rules for predicting gene function by examining the emergent structure and relationships between the gene functions and phenotypes rather than inspecting the terms semantically. The algorithms inspect relationships among multiple phenotype terms to deduce if there are cases where they all arise from a single gene function. We apply this methodology to data about genes in the laboratory mouse that are formally represented in the Mouse Genome Informatics (MGI) resource. From the data, 7444 rule instances were generated from five generalized rules, resulting in 4818 unique GO functional predictions for 1796 genes.
Conclusions: We show that our method is capable of inferring high-quality functional annotations from curated phenotype data. As well as creating inferred annotations, our method has the potential to allow for the elucidation of unforeseen, biologically significant associations between gene function and phenotypes that would be overlooked by a semantics-based approach. Future work will include the implementation of the described algorithms for a variety of other model organism databases, taking full advantage of the abundance of available high quality curated data.
Figures
References
-
- Gruber TR. A Translation Approach to Portable Ontology Specifications. Knowledge Acquisition. 1993;5(2):199–220. doi: 10.1006/knac.1993.1008. - DOI
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

