Extensive duplication of the Wolbachia DNA in chromosome four of Drosophila ananassae
- PMID: 25496002
- PMCID: PMC4299567
- DOI: 10.1186/1471-2164-15-1097
Extensive duplication of the Wolbachia DNA in chromosome four of Drosophila ananassae
Abstract
Background: Lateral gene transfer (LGT) from bacterial Wolbachia endosymbionts has been detected in ~20% of arthropod and nematode genome sequencing projects. Many of these transfers are large and contain a substantial part of the Wolbachia genome.
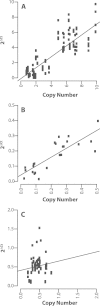
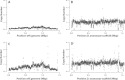
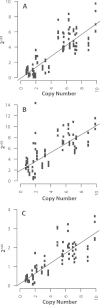
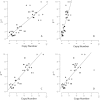
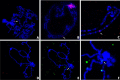
Results: Here, we re-sequenced three D. ananassae genomes from Asia and the Pacific that contain large LGTs from Wolbachia. We find that multiple copies of the Wolbachia genome are transferred to the Drosophila nuclear genome in all three lines. In the D. ananassae line from Indonesia, the copies of Wolbachia DNA in the nuclear genome are nearly identical in size and sequence yielding an even coverage of mapped reads over the Wolbachia genome. In contrast, the D. ananassae lines from Hawaii and India show an uneven coverage of mapped reads over the Wolbachia genome suggesting that different parts of these LGTs are present in different copy numbers. In the Hawaii line, we find that this LGT is underrepresented in third instar larvae indicative of being heterochromatic. Fluorescence in situ hybridization of mitotic chromosomes confirms that the LGT in the Hawaii line is heterochromatic and represents ~20% of the sequence on chromosome 4 (dot chromosome, Muller element F).
Conclusions: This collection of related lines contain large lateral gene transfers composed of multiple Wolbachia genomes that constitute >2% of the D. ananassae genome (~5 Mbp) and partially explain the abnormally large size of chromosome 4 in D. ananassae.
Figures
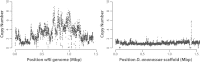

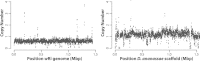

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

