Autonomous metabolomics for rapid metabolite identification in global profiling
- PMID: 25496351
- PMCID: PMC4303330
- DOI: 10.1021/ac5025649
Autonomous metabolomics for rapid metabolite identification in global profiling
Abstract
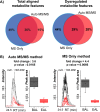
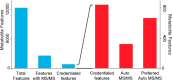
An autonomous metabolomic workflow combining mass spectrometry analysis with tandem mass spectrometry data acquisition was designed to allow for simultaneous data processing and metabolite characterization. Although previously tandem mass spectrometry data have been generated on the fly, the experiments described herein combine this technology with the bioinformatic resources of XCMS and METLIN. As a result of this unique integration, we can analyze large profiling datasets and simultaneously obtain structural identifications. Validation of the workflow on bacterial samples allowed the profiling on the order of a thousand metabolite features with simultaneous tandem mass spectra data acquisition. The tandem mass spectrometry data acquisition enabled automatic search and matching against the METLIN tandem mass spectrometry database, shortening the current workflow from days to hours. Overall, the autonomous approach to untargeted metabolomics provides an efficient means of metabolomic profiling, and will ultimately allow the more rapid integration of comparative analyses, metabolite identification, and data analysis at a systems biology level.
Figures



References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

