Molecular signatures that correlate with induction of lens regeneration in newts: lessons from proteomic analysis
- PMID: 25496664
- PMCID: PMC4271507
- DOI: 10.1186/s40246-014-0022-y
Molecular signatures that correlate with induction of lens regeneration in newts: lessons from proteomic analysis
Abstract
Background: Amphibians have the remarkable ability to regenerate missing body parts. After complete removal of the eye lens, the dorsal but not the ventral iris will transdifferentiate to regenerate an exact replica of the lost lens. We used reverse-phase nano-liquid chromatography followed by mass spectrometry to detect protein concentrations in dorsal and ventral iris 0, 4, and 8 days post-lentectomy. We performed gene expression comparisons between regeneration and intact timepoints as well as between dorsal and ventral iris.
Results: Our analysis revealed gene expression patterns associated with the ability of the dorsal iris for transdifferentiation and lens regeneration. Proteins regulating gene expression and various metabolic processes were enriched in regeneration timepoints. Proteins involved in extracellular matrix, gene expression, and DNA-associated functions like DNA repair formed a regeneration-related protein network and were all up-regulated in the dorsal iris. In addition, we investigated protein concentrations in cultured dorsal (transdifferentiation-competent) and ventral (transdifferentiation-incompetent) iris pigmented epithelial (IPE) cells. Our comparative analysis revealed that the ability of dorsal IPE cells to keep memory of their tissue of origin and transdifferentiation is associated with the expression of proteins that specify the dorso-ventral axis of the eye as well as with proteins found highly expressed in regeneration timepoints, especially 8 days post-lentectomy.
Conclusions: The study deepens our understanding in the mechanism of regeneration by providing protein networks and pathways that participate in the process.
Figures
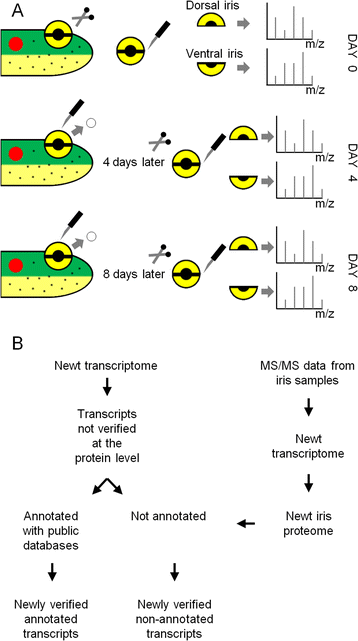
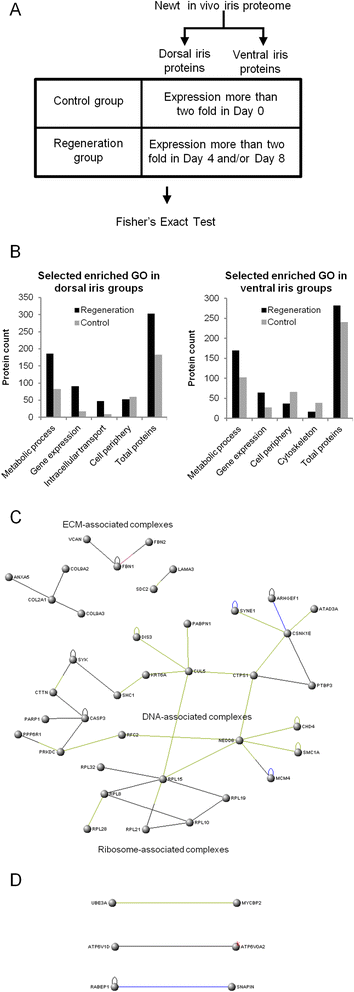
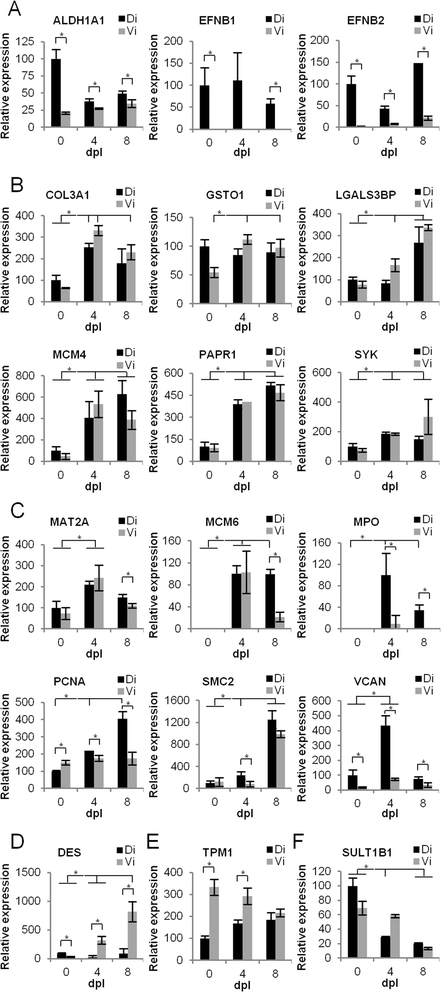


References
-
- Wolff G. Entwickelungsphysiologische studien. I. Die regeneration der urodelenlinse. Arch EntwMech Org. 1895;1:380–390.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

