Genomic signatures of near-extinction and rebirth of the crested ibis and other endangered bird species
- PMID: 25496777
- PMCID: PMC4290368
- DOI: 10.1186/s13059-014-0557-1
Genomic signatures of near-extinction and rebirth of the crested ibis and other endangered bird species
Abstract
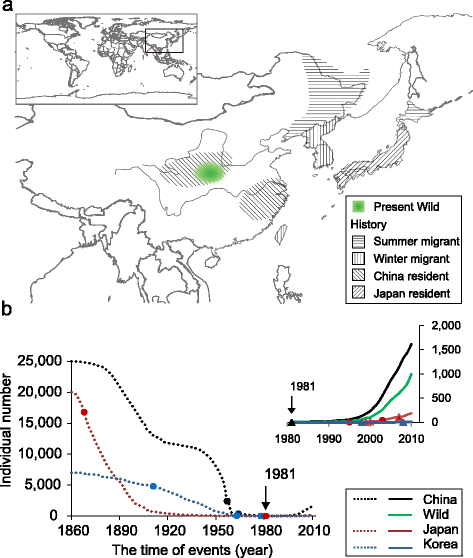
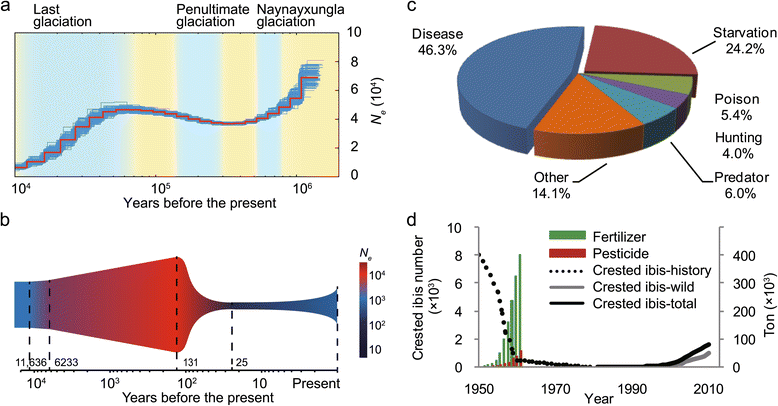
Background: Nearly one-quarter of all avian species is either threatened or nearly threatened. Of these, 73 species are currently being rescued from going extinct in wildlife sanctuaries. One of the previously most critically-endangered is the crested ibis, Nipponia nippon. Once widespread across North-East Asia, by 1981 only seven individuals from two breeding pairs remained in the wild. The recovering crested ibis populations thus provide an excellent example for conservation genomics since every individual bird has been recruited for genomic and demographic studies.
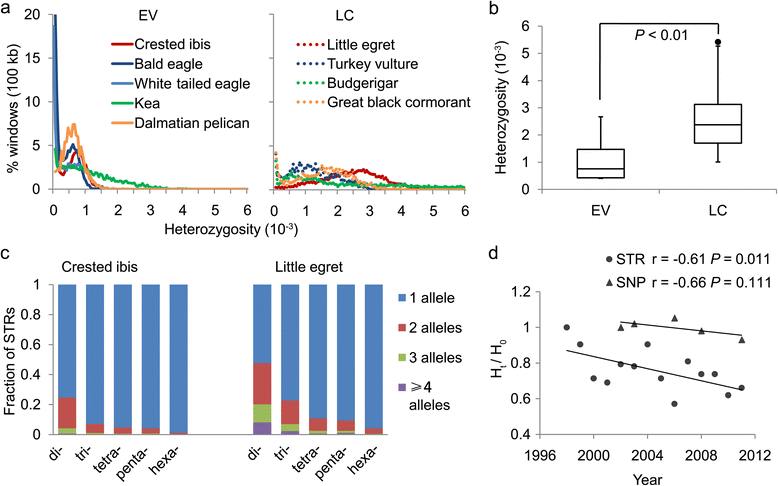
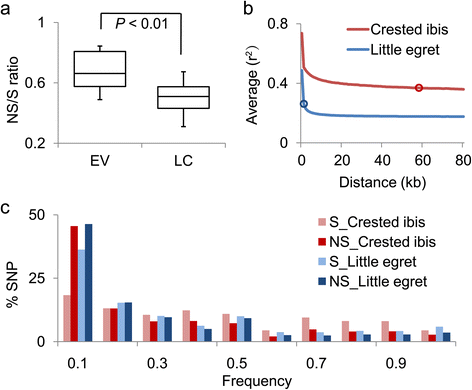
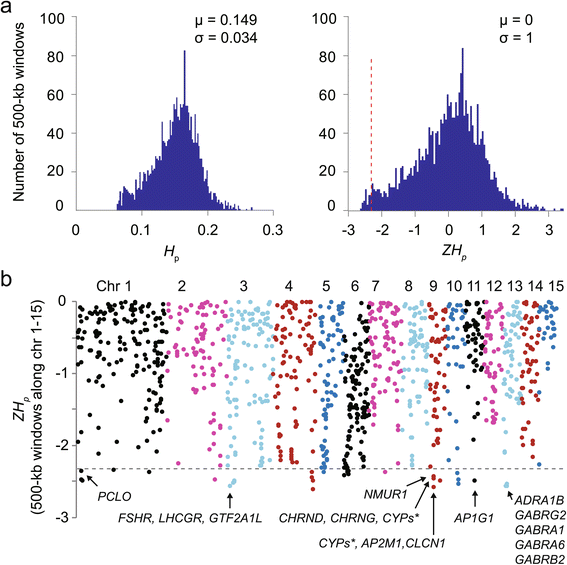
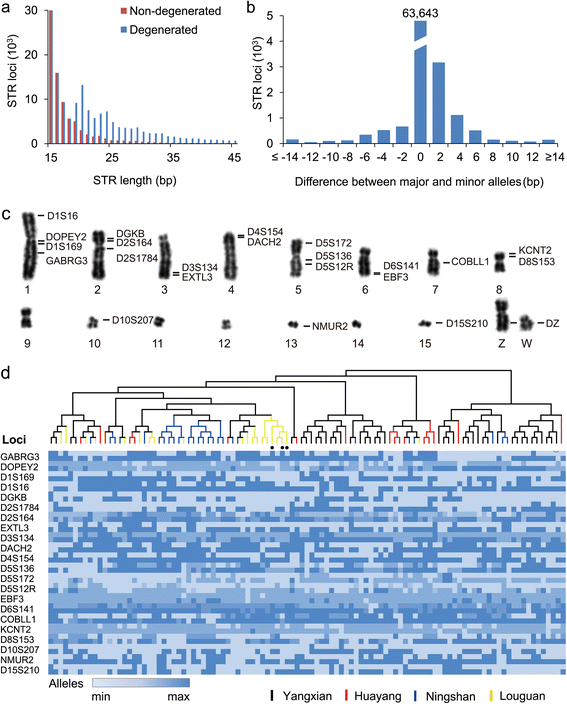
Results: Using high-quality genome sequences of multiple crested ibis individuals, its thriving co-habitant, the little egret, Egretta garzetta, and the recently sequenced genomes of 41 other avian species that are under various degrees of survival threats, including the bald eagle, we carry out comparative analyses for genomic signatures of near extinction events in association with environmental and behavioral attributes of species. We confirm that both loss of genetic diversity and enrichment of deleterious mutations of protein-coding genes contribute to the major genetic defects of the endangered species. We further identify that genetic inbreeding and loss-of-function genes in the crested ibis may all constitute genetic susceptibility to other factors including long-term climate change, over-hunting, and agrochemical overuse. We also establish a genome-wide DNA identification platform for molecular breeding and conservation practices, to facilitate sustainable recovery of endangered species.
Conclusions: These findings demonstrate common genomic signatures of population decline across avian species and pave a way for further effort in saving endangered species and enhancing conservation genomic efforts.
Figures
References
-
- Attenborough D. Help us build a brighter future: the need to save the world’s most threatened birds. Cambridge: BirdLife International; 2008.
-
- Collar NJ, Andreev AV, Chan S. Threatened Birds of Asia: The BirdLife International Red Data Book. 5. Cambridge: BirdLife International; 2001.
-
- Frankham R. Genetics and extinction. Biol Conserv. 2005;126:131–140. doi: 10.1016/j.biocon.2005.05.002. - DOI
-
- Saccheri I, Kuussaari M, Kankare M, Vikman P, Fortelius W, Hanski I. Inbreeding and extinction in a butterfly metapopulation. Nature. 1998;392:491–494. doi: 10.1038/33136. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

