The fundamental tradeoff in genomes and proteomes of prokaryotes established by the genetic code, codon entropy, and physics of nucleic acids and proteins
- PMID: 25496919
- PMCID: PMC4273451
- DOI: 10.1186/s13062-014-0029-2
The fundamental tradeoff in genomes and proteomes of prokaryotes established by the genetic code, codon entropy, and physics of nucleic acids and proteins
Abstract
Background: Mutations in nucleotide sequences provide a foundation for genetic variability, and selection is the driving force of the evolution and molecular adaptation. Despite considerable progress in the understanding of selective forces and their compositional determinants, the very nature of underlying mutational biases remains unclear.
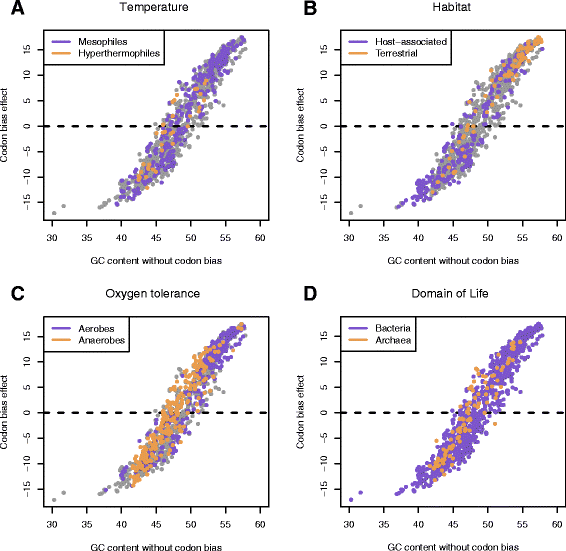
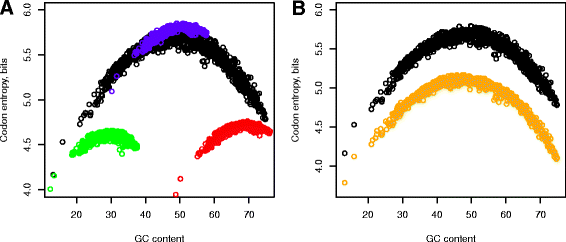
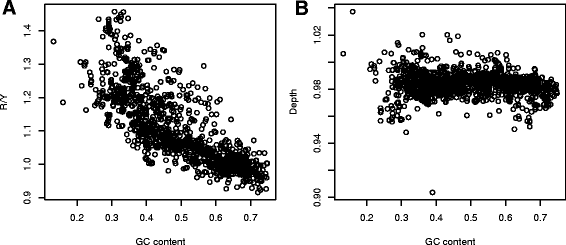
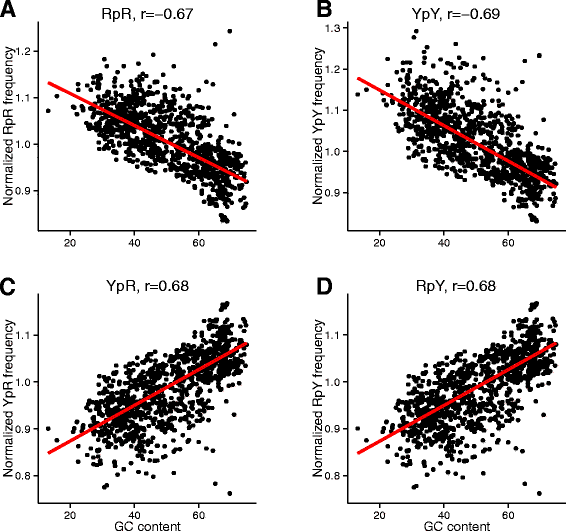
Results: We explore here a fundamental tradeoff, which analytically describes mutual adjustment of the nucleotide and amino acid compositions and its possible effect on the mutational biases. The tradeoff is determined by the interplay between the genetic code, optimization of the codon entropy, and demands on the structure and stability of nucleic acids and proteins.
Conclusion: The tradeoff is the unifying property of all prokaryotes regardless of the differences in their phylogenies, life styles, and extreme environments. It underlies mutational biases characteristic for genomes with different nucleotide and amino acid compositions, providing foundation for evolution and adaptation.
Figures



Similar articles
-
Thermophilic prokaryotes have characteristic patterns of codon usage, amino acid composition and nucleotide content.Gene. 2003 Oct 23;317(1-2):39-47. doi: 10.1016/s0378-1119(03)00660-7. Gene. 2003. PMID: 14604790
-
GC content-independent amino acid patterns in bacteria and archaea.J Basic Microbiol. 2012 Apr;52(2):195-205. doi: 10.1002/jobm.201100067. Epub 2011 Jul 21. J Basic Microbiol. 2012. PMID: 21780150
-
Evolution of proteomes: fundamental signatures and global trends in amino acid compositions.BMC Genomics. 2006 Dec 5;7:307. doi: 10.1186/1471-2164-7-307. BMC Genomics. 2006. PMID: 17147802 Free PMC article.
-
Driving change: the evolution of alternative genetic codes.Trends Genet. 2004 Feb;20(2):95-102. doi: 10.1016/j.tig.2003.12.009. Trends Genet. 2004. PMID: 14746991 Review.
-
Pathways of Genetic Code Evolution in Ancient and Modern Organisms.J Mol Evol. 2015 Jun;80(5-6):229-43. doi: 10.1007/s00239-015-9686-8. Epub 2015 Jun 9. J Mol Evol. 2015. PMID: 26054480 Review.
Cited by
-
Physicochemical Foundations of Life that Direct Evolution: Chance and Natural Selection are not Evolutionary Driving Forces.Life (Basel). 2020 Jan 21;10(2):7. doi: 10.3390/life10020007. Life (Basel). 2020. PMID: 31973071 Free PMC article.
-
The Mutational Robustness of the Genetic Code and Codon Usage in Environmental Context: A Non-Extremophilic Preference?Life (Basel). 2021 Jul 30;11(8):773. doi: 10.3390/life11080773. Life (Basel). 2021. PMID: 34440517 Free PMC article.
-
No evidence for widespread positive selection on double substitutions within codons in primates and yeasts.Front Genet. 2022 Sep 9;13:991249. doi: 10.3389/fgene.2022.991249. eCollection 2022. Front Genet. 2022. PMID: 36159983 Free PMC article.
-
A proteome-scale analysis of vertebrate protein amino acid occurrence: Thermoadaptation shows a correlation with protein solvation but less so with dynamics.Proteins. 2023 Jan;91(1):3-15. doi: 10.1002/prot.26404. Epub 2022 Aug 20. Proteins. 2023. PMID: 36053994 Free PMC article.
-
Deciphering the rationale behind specific codon usage pattern in extremophiles.Sci Rep. 2018 Oct 19;8(1):15548. doi: 10.1038/s41598-018-33476-x. Sci Rep. 2018. PMID: 30341344 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

