Comparing viral metagenomics methods using a highly multiplexed human viral pathogens reagent
- PMID: 25497414
- PMCID: PMC4344864
- DOI: 10.1016/j.jviromet.2014.12.002
Comparing viral metagenomics methods using a highly multiplexed human viral pathogens reagent
Abstract
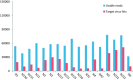
Unbiased metagenomic sequencing holds significant potential as a diagnostic tool for the simultaneous detection of any previously genetically described viral nucleic acids in clinical samples. Viral genome sequences can also inform on likely phenotypes including drug susceptibility or neutralization serotypes. In this study, different variables of the laboratory methods often used to generate viral metagenomics libraries were compared for their abilities to detect multiple viruses and generate full genome coverage. A biological reagent consisting of 25 different human RNA and DNA viral pathogens was used to estimate the effect of filtration and nuclease digestion, DNA/RNA extraction methods, pre-amplification and the use of different library preparation kits on the detection of viral nucleic acids. Filtration and nuclease treatment led to slight decreases in the percentage of viral sequence reads and number of viruses detected. For nucleic acid extractions silica spin columns improved viral sequence recovery relative to magnetic beads and Trizol extraction. Pre-amplification using random RT-PCR while generating more viral sequence reads resulted in detection of fewer viruses, more overlapping sequences, and lower genome coverage. The ScriptSeq library preparation method retrieved more viruses and a greater fraction of their genomes than the TruSeq and Nextera methods. Viral metagenomics sequencing was able to simultaneously detect up to 22 different viruses in the biological reagent analyzed including all those detected by qPCR. Further optimization will be required for the detection of viruses in biologically more complex samples such as tissues, blood, or feces.
Keywords: Diagnosis; Method; Next generation sequencing; Viral metagenomics; Virus pathogen discovery.
Copyright © 2014 Elsevier B.V. All rights reserved.
Figures


References
-
- Bidzhieva B., Zagorodnyaya T., Karagiannis K., Simonyan V., Laassri M., Chumakov K. Deep sequencing approach for genetic stability evaluation of influenza A viruses. J. Virol. Methods. 2014;199:68–75. - PubMed
-
- Capobianchi M.R., Giombini E., Rozera G. Next-generation sequencing technology in clinical virology. Clin. Microbiol. Infect. 2013;19:15–22. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

