Evolution and gene capture in ancient endogenous retroviruses - insights from the crocodilian genomes
- PMID: 25499090
- PMCID: PMC4299795
- DOI: 10.1186/s12977-014-0071-2
Evolution and gene capture in ancient endogenous retroviruses - insights from the crocodilian genomes
Abstract
Background: Crocodilians are thought to be hosts to a diverse and divergent complement of endogenous retroviruses (ERVs) but a comprehensive investigation is yet to be performed. The recent sequencing of three crocodilian genomes provides an opportunity for a more detailed and accurate representation of the ERV diversity that is present in these species. Here we investigate the diversity, distribution and evolution of ERVs from the genomes of three key crocodilian species, and outline the key processes driving crocodilian ERV proliferation and evolution.
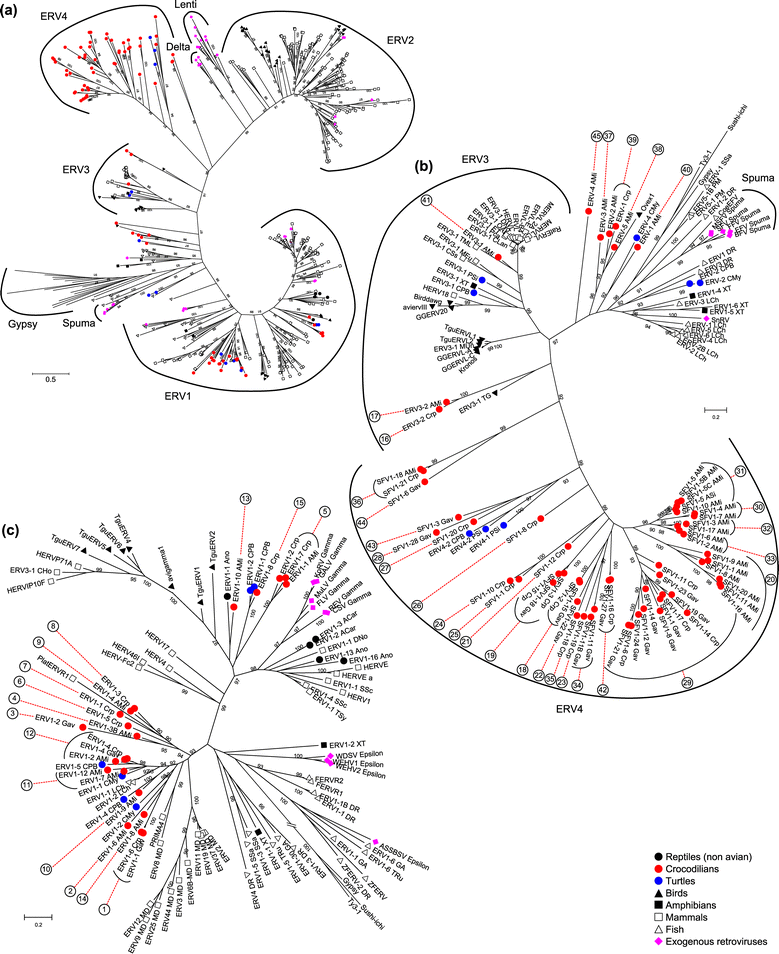
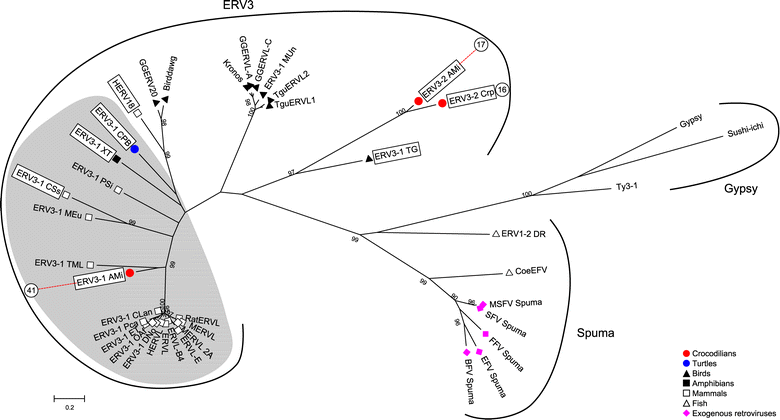
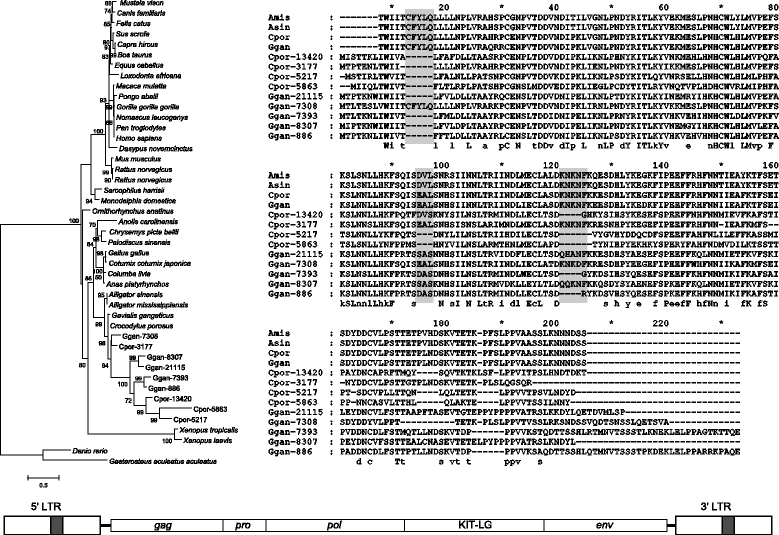
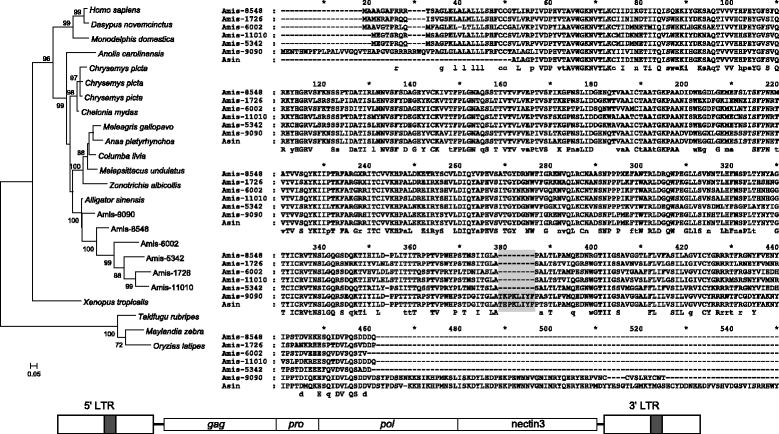
Results: ERVs and ERV related sequences make up less than 2% of crocodilian genomes. We recovered and described 45 ERV groups within the three crocodilian genomes, many of which are species specific. We have also revealed a new class of ERV, ERV4, which appears to be common to crocodilians and turtles, and currently has no characterised exogenous counterpart. For the first time, we formally describe the characteristics of this ERV class and its classification relative to other recognised ERV and retroviral classes. This class shares some sequence similarity and sequence characteristics with ERV3, although it is phylogenetically distinct from the other ERV classes. We have also identified two instances of gene capture by crocodilian ERVs, one of which, the capture of a host KIT-ligand mRNA has occurred without the loss of an ERV domain.
Conclusions: This study indicates that crocodilian ERVs comprise a wide variety of lineages, many of which appear to reflect ancient infections. In particular, ERV4 appears to have a limited host range, with current data suggesting that it is confined to crocodilians and some lineages of turtles. Also of interest are two ERV groups that demonstrate evidence of host gene capture. This study provides a framework to facilitate further studies into non-mammalian vertebrates and highlights the need for further studies into such species.
Figures
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

