Using the class 1 integron-integrase gene as a proxy for anthropogenic pollution
- PMID: 25500508
- PMCID: PMC4438328
- DOI: 10.1038/ismej.2014.226
Using the class 1 integron-integrase gene as a proxy for anthropogenic pollution
Abstract
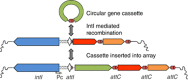
Around all human activity, there are zones of pollution with pesticides, heavy metals, pharmaceuticals, personal care products and the microorganisms associated with human waste streams and agriculture. This diversity of pollutants, whose concentration varies spatially and temporally, is a major challenge for monitoring. Here, we suggest that the relative abundance of the clinical class 1 integron-integrase gene, intI1, is a good proxy for pollution because: (1) intI1 is linked to genes conferring resistance to antibiotics, disinfectants and heavy metals; (2) it is found in a wide variety of pathogenic and nonpathogenic bacteria; (3) its abundance can change rapidly because its host cells can have rapid generation times and it can move between bacteria by horizontal gene transfer; and (4) a single DNA sequence variant of intI1 is now found on a wide diversity of xenogenetic elements, these being complex mosaic DNA elements fixed through the agency of human selection. Here we review the literature examining the relationship between anthropogenic impacts and the abundance of intI1, and outline an approach by which intI1 could serve as a proxy for anthropogenic pollution.
Figures

References
-
- Bailey JK, Pinyon JL, Anantham S, Hall RM. Commensal Escherichia coli of healthy humans: a reservoir for antibiotic-resistance determinants. J Med Microbiol. 2010;59:1331–1339. - PubMed
-
- Baker-Austin C, Wright MS, Stepanauskas R, McArthur JV. Co-selection of antibiotic and metal resistance. Trends Microbiol. 2006;14:176–182. - PubMed
-
- Berglund B, Khan GA, Weisner SEB, Ehde PM, Fick J, Lindgren P-E. Efficient removal of antibiotics in surface-flow constructed wetlands, with no observed impact on antibiotic resistance genes. Sci Total Environ. 2014;476–477:29–37. - PubMed
-
- Binh CTT, Heuer H, Kaupenjohann M, Smalla K. Diverse aadA gene cassettes on class 1 integrons introduced into soil via spread manure. Res Microbiol. 2009;160:427–433. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

