A structural perspective on nuclear receptors as targets of environmental compounds
- PMID: 25500867
- PMCID: PMC4571321
- DOI: 10.1038/aps.2014.133
A structural perspective on nuclear receptors as targets of environmental compounds
Abstract
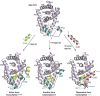
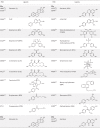
Nuclear receptors (NRs) are members of a large superfamily of evolutionarily related transcription factors that control a plethora of biological processes. NRs orchestrate complex events such as development, organ homeostasis, metabolism, immune function, and reproduction. Approximately one-half of the 48 human NRs have been shown to act as ligand-regulated transcription factors and respond directly to a large variety of endogenous hormones and metabolites that are generally hydrophobic and small in size (eg, retinoic acid or estradiol). The second half of the NR family comprises the so-called orphan receptors, for which regulatory ligands are still unknown or may not exist despite the presence of a C-terminal ligand-binding domain, which is the hallmark of all NRs. Several chemicals released into the environment (eg, bisphenols, phthalates, parabens, etc) share some physicochemical properties with natural ligands, allowing them to bind to NRs and activate or inhibit their action. Collectively referred to as endocrine disruptors or endocrine-disrupting chemicals (EDCs), these environmental pollutants are highly suspected to cause a wide range of developmental, reproductive, neurological, or metabolic defects in humans and wildlife. Crystallographic studies are revealing unanticipated mechanisms by which chemically diverse EDCs interact with the ligand-binding domain of NRs. These studies thereby provide a rational basis for designing novel chemicals with lower impacts on human and animal health. In this review, we provide a structural and mechanistic view of endocrine disrupting action using estrogen receptors α and β, (ERα/β), peroxisome proliferator activated receptor γ (PPARγ), and their respective environmental ligands as representative examples.
Figures

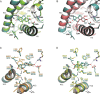

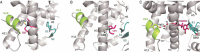

References
-
- Germain P, Staels B, Dacquet C, Spedding M, Laudet V. Overview of nomenclature of nuclear receptors. Pharmacol Rev 2006; 58: 685–704. - PubMed
-
- Gronemeyer H, Gustafsson JA, Laudet V. Principles for modulation of the nuclear receptor superfamily. Nat Rev Drug Discov 2004; 3: 950–64. - PubMed
-
- Li Y, Lambert MH, Xu HE. Activation of nuclear receptors: a perspective from structural genomics. Structure 2003; 11: 741–6. - PubMed
-
- Bourguet W, Germain P, Gronemeyer H. Nuclear receptor ligand-binding domains: three-dimensional structures, molecular interactions and pharmacological implications. Trends Pharmacol Sci 2000; 21: 381–8. - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

