A region-specific neurogenesis mode requires migratory progenitors in the Drosophila visual system
- PMID: 25501037
- PMCID: PMC4338547
- DOI: 10.1038/nn.3896
A region-specific neurogenesis mode requires migratory progenitors in the Drosophila visual system
Erratum in
-
Erratum: A region-specific neurogenesis mode requires migratory progenitors in the Drosophila visual system.Nat Neurosci. 2015 Jun;18(6):926. doi: 10.1038/nn0615-926b. Nat Neurosci. 2015. PMID: 26007215 No abstract available.
Abstract
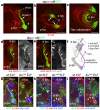
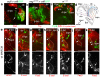
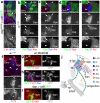
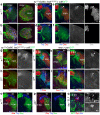
Brain areas each generate specific neuron subtypes during development. However, underlying regional variations in neurogenesis strategies and regulatory mechanisms remain poorly understood. In Drosophila, neurons in four optic lobe ganglia originate from two neuroepithelia, the outer (OPC) and inner (IPC) proliferation centers. Using genetic manipulations, we found that one IPC neuroepithelial domain progressively transformed into migratory progenitors that matured into neural stem cells (neuroblasts) in a second domain. Progenitors emerged by an epithelial-mesenchymal transition-like mechanism that required the Snail-family member Escargot and, in subdomains, Decapentaplegic signaling. The proneural factors Lethal of scute and Asense differentially controlled progenitor supply and maturation into neuroblasts. These switched expression from Asense to a third proneural protein, Atonal. Dichaete and Tailless mediated this transition, which was essential for generating two neuron populations at defined positions. We propose that this neurogenesis mode is central for setting up a new proliferative zone to facilitate spatio-temporal matching of neurogenesis and connectivity across ganglia.
Figures




References
-
- Florio M, Huttner WB. Neural progenitors, neurogenesis and the evolution of the neocortex. Development. 2014;141:2182–2194. - PubMed
-
- Brand AH, Livesey FJ. Neural stem cell biology in vertebrates and invertebrates: more alike than different? Neuron. 2011;70:719–729. - PubMed
-
- Homem CC, Knoblich JA. Drosophila neuroblasts: a model for stem cell biology. Development. 2012;139:4297–4310. - PubMed
-
- Ayala R, Shu T, Tsai LH. Trekking across the brain: the journey of neuronal migration. Cell. 2007;128:29–43. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

