Emergence shapes the structure of the seed microbiota
- PMID: 25501471
- PMCID: PMC4309697
- DOI: 10.1128/AEM.03722-14
Emergence shapes the structure of the seed microbiota
Abstract
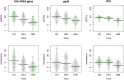
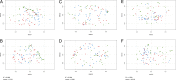
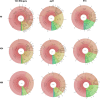
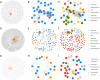
Seeds carry complex microbial communities, which may exert beneficial or deleterious effects on plant growth and plant health. To date, the composition of microbial communities associated with seeds has been explored mainly through culture-based diversity studies and therefore remains largely unknown. In this work, we analyzed the structures of the seed microbiotas of different plants from the family Brassicaceae and their dynamics during germination and emergence through sequencing of three molecular markers: the ITS1 region of the fungal internal transcribed spacer, the V4 region of 16S rRNA gene, and a species-specific bacterial marker based on a fragment of gyrB. Sequence analyses revealed important variations in microbial community composition between seed samples. Moreover, we found that emergence strongly influences the structure of the microbiota, with a marked reduction of bacterial and fungal diversity. This shift in the microbial community composition is mostly due to an increase in the relative abundance of some bacterial and fungal taxa possessing fast-growing abilities. Altogether, our results provide an estimation of the role of the seed as a source of inoculum for the seedling, which is crucial for practical applications in developing new strategies of inoculation for disease prevention.
Figures
References
-
- Baker KF, Smith SH. 1966. Dynamics of seed transmission of plant pathogens. Annu Rev Phytopathol 4:311–332. doi: 10.1146/annurev.py.04.090166.001523. - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

