Role of plasmids in Lactobacillus brevis BSO 464 hop tolerance and beer spoilage
- PMID: 25501474
- PMCID: PMC4309717
- DOI: 10.1128/AEM.02870-14
Role of plasmids in Lactobacillus brevis BSO 464 hop tolerance and beer spoilage
Abstract
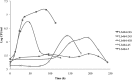
Specific isolates of lactic acid bacteria (LAB) can grow in the harsh beer environment, thus posing a threat to brew quality and the economic success of breweries worldwide. Plasmid-localized genes, such as horA, horC, and hitA, have been suggested to confer hop tolerance, a trait required for LAB survival in beer. The presence and expression of these genes among LAB, however, do not universally correlate with the ability to grow in beer. Genome sequencing of the virulent beer spoilage organism Lactobacillus brevis BSO 464 revealed the presence of eight plasmids, with plasmids 1, 2, and 3 containing horA, horC, and hitA, respectively. To investigate the roles that these and the other five plasmids play in L. brevis BSO 464 growth in beer, plasmid curing with novobiocin was used to derive 10 plasmid variants. Multiplex PCRs were utilized to determine the presence or absence of each plasmid, and how plasmid loss affected hop tolerance and growth in degassed (noncarbonated) beer was assessed. Loss of three of the eight plasmids was found to affect hop tolerance and growth in beer. Loss of plasmid 2 (horC and 28 other genes) had the most dramatic effect, with loss of plasmid 4 (120 genes) and plasmid 8 (47 genes) having significant, but smaller, impacts. These results support the contention that genes on mobile genetic elements are essential for bacterial growth in beer and that beer spoilage ability is not dependent solely on the three previously described hop tolerance genes or on the chromosome of a beer spoilage LAB isolate.
Figures

References
-
- Simpson WJ, Fernandez JL. 1992. Selection of beer-spoilage lactic acid bacteria and induction of their ability to grow in beer. Lett Appl Microbiol 14:13–16. doi:10.1111/j.1472-765X.1992.tb00636.x. - DOI
-
- Bamforth CW. 2006. Scientific principles of malting and brewing. American Society of Brewing Chemists, St. Paul, MN.
-
- Priest FG, Campbell I. 2003. Brewing microbiology. Kluwer Academic, New York, NY.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

