Steady-state kinetic analysis of DNA polymerase single-nucleotide incorporation products
- PMID: 25501593
- PMCID: PMC4274652
- DOI: 10.1002/0471142700.nc0721s59
Steady-state kinetic analysis of DNA polymerase single-nucleotide incorporation products
Abstract
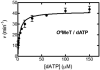
This unit describes the experimental procedures for the steady-state kinetic analysis of DNA synthesis across DNA nucleotides (native or modified) by DNA polymerases. In vitro primer extension experiments with a single nucleoside triphosphate species followed by denaturing polyacrylamide gel electrophoresis of the extended products is described. Data analysis procedures and fitting to steady-state kinetic models is presented to highlight the kinetic differences involved in the bypass of damaged versus undamaged DNA. Moreover, explanations concerning problems encountered in these experiments are addressed. This approach provides useful quantitative parameters for the processing of damaged DNA by DNA polymerases.
Keywords: DNA polymerase; steady-state kinetics; translesion synthesis.
Copyright © 2014 John Wiley & Sons, Inc.
Figures



References
-
- Berg JM, Tymoczko JL, Stryer L. Enzymes: Basic concepts and kinetics. In: Samols L, Hadler GL, Zimmerman P, Brooks N, editors. Biochemistry. Seventh Ed. W. H. Freeman; New York: 2012. pp. 229–237.
-
- Boosalis MS, Petruska J, Goodman MF. DNA polymerase insertion fidelity: gel assay for site-specific kinetics. J. Biol. Chem. 1987;262:14689–96. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

