A new class of highly potent, broadly neutralizing antibodies isolated from viremic patients infected with dengue virus
- PMID: 25501631
- PMCID: PMC4445969
- DOI: 10.1038/ni.3058
A new class of highly potent, broadly neutralizing antibodies isolated from viremic patients infected with dengue virus
Erratum in
-
Corrigendum: A new class of highly potent, broadly neutralizing antibodies isolated from viremic patients infected with dengue virus.Nat Immunol. 2015 May;16(5):544. doi: 10.1038/ni0515-544b. Nat Immunol. 2015. PMID: 25898199 No abstract available.
-
Corrigendum: A new class of highly potent, broadly neutralizing antibodies isolated from viremic patients infected with dengue virus.Nat Immunol. 2015 Jul;16(7):785. doi: 10.1038/ni0715-785a. Nat Immunol. 2015. PMID: 26086146 No abstract available.
Abstract
Dengue is a rapidly emerging, mosquito-borne viral infection, with an estimated 400 million infections occurring annually. To gain insight into dengue immunity, we characterized 145 human monoclonal antibodies (mAbs) and identified a previously unknown epitope, the envelope dimer epitope (EDE), that bridges two envelope protein subunits that make up the 90 repeating dimers on the mature virion. The mAbs to EDE were broadly reactive across the dengue serocomplex and fully neutralized virus produced in either insect cells or primary human cells, with 50% neutralization in the low picomolar range. Our results provide a path to a subunit vaccine against dengue virus and have implications for the design and monitoring of future vaccine trials in which the induction of antibody to the EDE should be prioritized.
Figures



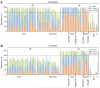



Comment in
-
Broad and strong: the ultimate antibody to dengue virus.Nat Immunol. 2015 Feb;16(2):135-7. doi: 10.1038/ni.3081. Nat Immunol. 2015. PMID: 25594456 No abstract available.
References
-
- Westaway EG, Blok J. In: Dengue and Dengue Hemorrhagic fever. Gubler DJ, Kuno G, editors. CABI Publishing; Oxford, UK: 1997. pp. 147–173.
-
- Mongkolsapaya J, et al. Original antigenic sin and apoptosis in the pathogenesis of dengue hemorrhagic fever. Nat. Med. 2003;9:921–927. - PubMed
-
- Mongkolsapaya J, et al. T cell responses in dengue hemorrhagic fever: are cross-reactive T cells suboptimal? J. Immunol. 2006;176:3821–3829. - PubMed
-
- Sangkawibha N, et al. Risk factors in dengue shock syndrome: a prospective epidemiologic study in Rayong, Thailand. I. The 1980 outbreak. Am. J. Epidemiol. 1984;120:653–669. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 1S10OD018111/OD/NIH HHS/United States
- 095541/WT_/Wellcome Trust/United Kingdom
- 089276/WT_/Wellcome Trust/United Kingdom
- S10 OD018111/OD/NIH HHS/United States
- G0801508/MRC_/Medical Research Council/United Kingdom
- AI094386/AI/NIAID NIH HHS/United States
- G0400720/MRC_/Medical Research Council/United Kingdom
- S10 RR023057/RR/NCRR NIH HHS/United States
- R01 AI094386/AI/NIAID NIH HHS/United States
- GM071940/GM/NIGMS NIH HHS/United States
- G0600000/MRC_/Medical Research Council/United Kingdom
- R01 GM071940/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

