An open source image processing method to quantitatively assess tissue growth after non-invasive magnetic resonance imaging in human bone marrow stromal cell seeded 3D polymeric scaffolds
- PMID: 25502022
- PMCID: PMC4264848
- DOI: 10.1371/journal.pone.0115000
An open source image processing method to quantitatively assess tissue growth after non-invasive magnetic resonance imaging in human bone marrow stromal cell seeded 3D polymeric scaffolds
Abstract
Monitoring extracellular matrix (ECM) components is one of the key methods used to determine tissue quality in three-dimensional (3D) scaffolds for regenerative medicine and clinical purposes. This is even more important when multipotent human bone marrow stromal cells (hMSCs) are used, as it could offer a method to understand in real time the dynamics of stromal cell differentiation and eventually steer it into the desired lineage. Magnetic Resonance Imaging (MRI) is a promising tool to overcome the challenge of a limited transparency in opaque 3D scaffolds. Technical limitations of MRI involve non-uniform background intensity leading to fluctuating background signals and therewith complicating quantifications on the retrieved images. We present a post-imaging processing sequence that is able to correct for this non-uniform background intensity. To test the processing sequence we investigated the use of MRI for in vitro monitoring of tissue growth in three-dimensional poly(ethylene oxide terephthalate)-poly(butylene terephthalate) (PEOT/PBT) scaffolds. Results showed that MRI, without the need to use contrast agents, is a promising non-invasive tool to quantitatively monitor ECM production and cell distribution during in vitro culture in 3D porous tissue engineered constructs.
Conflict of interest statement
Figures
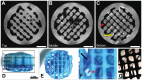
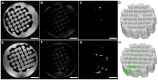


Similar articles
-
Label-free Raman monitoring of extracellular matrix formation in three-dimensional polymeric scaffolds.J R Soc Interface. 2013 Jul 3;10(86):20130464. doi: 10.1098/rsif.2013.0464. Print 2013 Sep 6. J R Soc Interface. 2013. PMID: 23825118 Free PMC article.
-
Biological and Tribological Assessment of Poly(Ethylene Oxide Terephthalate)/Poly(Butylene Terephthalate), Polycaprolactone, and Poly (L\DL) Lactic Acid Plotted Scaffolds for Skeletal Tissue Regeneration.Adv Healthc Mater. 2016 Jan 21;5(2):232-43. doi: 10.1002/adhm.201500067. Epub 2015 Nov 25. Adv Healthc Mater. 2016. PMID: 26775915
-
Combining technologies to create bioactive hybrid scaffolds for bone tissue engineering.Biomatter. 2013 Apr-Jun;3(2):e23705. doi: 10.4161/biom.23705. Epub 2013 Jan 1. Biomatter. 2013. PMID: 23507924 Free PMC article.
-
Hyaluronic Acid (HA) Scaffolds and Multipotent Stromal Cells (MSCs) in Regenerative Medicine.Stem Cell Rev Rep. 2016 Dec;12(6):664-681. doi: 10.1007/s12015-016-9684-2. Stem Cell Rev Rep. 2016. PMID: 27665291 Review.
-
Recent Advances in Endocrine, Metabolic and Immune Disorders: Mesenchymal Stem Cells (MSCs) and Engineered Scaffolds.Endocr Metab Immune Disord Drug Targets. 2018;18(5):466-469. doi: 10.2174/1871530318666180423102905. Endocr Metab Immune Disord Drug Targets. 2018. PMID: 29692270 Review.
Cited by
-
Towards MRI Study of Biointegration of Carbon-Carbon Composites with Ca-P Coatings.Nanomaterials (Basel). 2025 Mar 26;15(7):492. doi: 10.3390/nano15070492. Nanomaterials (Basel). 2025. PMID: 40214538 Free PMC article.
-
3D-printed bioactive scaffolds from nanosilicates and PEOT/PBT for bone tissue engineering.Regen Biomater. 2019 Feb;6(1):29-37. doi: 10.1093/rb/rby024. Epub 2018 Dec 15. Regen Biomater. 2019. PMID: 30740240 Free PMC article.
References
-
- Chen WL, Huang CH, Chiou LL, Chen TH, Huang YY, et al. (2010) Multiphoton imaging and quantitative analysis of collagen production by chondrogenic human mesenchymal stem cells cultured in chitosan scaffold. Tissue Eng Part C Methods 16:913–920. - PubMed
-
- Washburn NR, Weir M, Anderson P, Potter K (2004) Bone formation in polymeric scaffolds evaluated by proton magnetic resonance microscopy and X-ray microtomography. Journal of Biomedical Materials Research Part A 69A:738–747. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

