Drug resistance. Population transcriptomics of human malaria parasites reveals the mechanism of artemisinin resistance
- PMID: 25502316
- PMCID: PMC5642863
- DOI: 10.1126/science.1260403
Drug resistance. Population transcriptomics of human malaria parasites reveals the mechanism of artemisinin resistance
Abstract
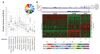
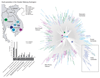
Artemisinin resistance in Plasmodium falciparum threatens global efforts to control and eliminate malaria. Polymorphisms in the kelch domain-carrying protein K13 are associated with artemisinin resistance, but the underlying molecular mechanisms are unknown. We analyzed the in vivo transcriptomes of 1043 P. falciparum isolates from patients with acute malaria and found that artemisinin resistance is associated with increased expression of unfolded protein response (UPR) pathways involving the major PROSC and TRiC chaperone complexes. Artemisinin-resistant parasites also exhibit decelerated progression through the first part of the asexual intraerythrocytic development cycle. These findings suggest that artemisinin-resistant parasites remain in a state of decelerated development at the young ring stage, whereas their up-regulated UPR pathways mitigate protein damage caused by artemisinin. The expression profiles of UPR-related genes also associate with the geographical origin of parasite isolates, further suggesting their role in emerging artemisinin resistance in the Greater Mekong Subregion.
Copyright © 2015, American Association for the Advancement of Science.
Figures

Comment in
-
Infectious diseases. Understanding artemisinin resistance.Science. 2015 Jan 23;347(6220):373-4. doi: 10.1126/science.aaa4102. Science. 2015. PMID: 25613877 No abstract available.
References
Publication types
MeSH terms
Substances
Supplementary concepts
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

