"Ménage à Trois": the evolutionary interplay between JSRV, enJSRVs and domestic sheep
- PMID: 25502326
- PMCID: PMC4276937
- DOI: 10.3390/v6124926
"Ménage à Trois": the evolutionary interplay between JSRV, enJSRVs and domestic sheep
Abstract
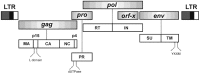
Sheep betaretroviruses represent a fascinating model to study the complex evolutionary interplay between host and pathogen in natural settings. In infected sheep, the exogenous and pathogenic Jaagsiekte sheep retrovirus (JSRV) coexists with a variety of highly related endogenous JSRVs, referred to as enJSRVs. During evolution, some of them were co-opted by the host as they fulfilled important biological functions, including placental development and protection against related exogenous retroviruses. In particular, two enJSRV loci, enJS56A1 and enJSRV-20, were positively selected during sheep domestication due to their ability to interfere with the replication of related competent retroviruses. Interestingly, viruses escaping these transdominant enJSRVs have recently emerged, probably less than 200 years ago. Overall, these findings suggest that in sheep the process of endogenization is still ongoing and, therefore, the evolutionary interplay between endogenous and exogenous sheep betaretroviruses and their host has not yet reached an equilibrium.
Figures



References
-
- Stoye J.P. Studies of endogenous retroviruses reveal a continuing evolutionary saga. Nat. Rev. Microbiol. 2012;10:395–406. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

