Chromosomal Instability and Phosphoinositide Pathway Gene Signatures in Glioblastoma Multiforme
- PMID: 25502460
- PMCID: PMC4703635
- DOI: 10.1007/s12035-014-9034-9
Chromosomal Instability and Phosphoinositide Pathway Gene Signatures in Glioblastoma Multiforme
Abstract
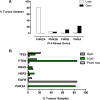
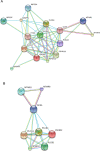
Structural rearrangements of chromosome 10 are frequently observed in glioblastoma multiforme and over 80 % of tumour samples archived in the catalogue of somatic mutations in cancer database had gene copy number loss for PI4K2A which encodes phosphatidylinositol 4-kinase type IIalpha. PI4K2A loss of heterozygosity mirrored that of PTEN, another enzyme that regulates phosphoinositide levels and also PIK3AP1, MINPP1, INPP5A and INPP5F. These results indicated a reduction in copy number for a set of phosphoinositide signalling genes that co-localise to chromosome 10q. This analysis was extended to a panel of phosphoinositide pathway genes on other chromosomes and revealed a number of previously unreported associations with glioblastoma multiforme. Of particular note were highly penetrant copy number losses for a group of X-linked phosphoinositide phosphatase genes OCRL, MTM1 and MTMR8; copy number amplifications for the chromosome 19 genes PIP5K1C, AKT2 and PIK3R2, and also for the phospholipase C genes PLCB1, PLCB4 and PLCG1 on chromosome 20. These mutations are likely to affect signalling and trafficking functions dependent on the PI(4,5)P2, PI(3,4,5)P3 and PI(3,5)P2 lipids as well as the inositol phosphates IP3, IP5 and IP6. Analysis of flanking genes with functionally unrelated products indicated that chromosomal instability as opposed to a phosphoinositide-specific process underlay this pattern of copy number variation. This in silico study suggests that in glioblastoma multiforme, karyotypic changes have the potential to cause multiple abnormalities in sets of genes involved in phosphoinositide metabolism and this may be important for understanding drug resistance and phosphoinositide pathway redundancy in the advanced disease state.
Keywords: Cancer; Gene copy number; Glioblastoma; PI 4-kinase.
Conflict of interest statement
The author declares no conflict of interest.
Figures

Similar articles
-
Phosphoinositide 5-phosphatase activities control cell motility in glioblastoma: Two phosphoinositides PI(4,5)P2 and PI(3,4)P2 are involved.Adv Biol Regul. 2018 Jan;67:40-48. doi: 10.1016/j.jbior.2017.09.001. Epub 2017 Sep 5. Adv Biol Regul. 2018. PMID: 28916189 Review.
-
Differential SKIP expression in PTEN-deficient glioblastoma regulates cellular proliferation and migration.Oncogene. 2015 Jul;34(28):3711-27. doi: 10.1038/onc.2014.303. Epub 2014 Sep 22. Oncogene. 2015. PMID: 25241900
-
High-resolution genomic copy number profiling of glioblastoma multiforme by single nucleotide polymorphism DNA microarray.Mol Cancer Res. 2009 May;7(5):665-77. doi: 10.1158/1541-7786.MCR-08-0270. Epub 2009 May 12. Mol Cancer Res. 2009. PMID: 19435819
-
High-resolution genome-wide allelotype analysis identifies loss of chromosome 14q as a recurrent genetic alteration in astrocytic tumours.Br J Cancer. 2002 Jul 15;87(2):218-24. doi: 10.1038/sj.bjc.6600430. Br J Cancer. 2002. PMID: 12107846 Free PMC article.
-
Genetic pathways to primary and secondary glioblastoma.Am J Pathol. 2007 May;170(5):1445-53. doi: 10.2353/ajpath.2007.070011. Am J Pathol. 2007. PMID: 17456751 Free PMC article. Review.
Cited by
-
U-73122 reduces the cell growth in cultured MG-63 ostesarcoma cell line involving Phosphoinositide-specific Phospholipases C.Springerplus. 2016 Feb 24;5:156. doi: 10.1186/s40064-016-1768-6. eCollection 2016. Springerplus. 2016. PMID: 27026853 Free PMC article.
-
MiR-1246 regulates the PI3K/AKT signaling pathway by targeting PIK3AP1 and inhibits thyroid cancer cell proliferation and tumor growth.Mol Cell Biochem. 2022 Mar;477(3):649-661. doi: 10.1007/s11010-021-04290-3. Epub 2021 Dec 6. Mol Cell Biochem. 2022. PMID: 34870753 Free PMC article.
-
Identification of key modules and hub genes in glioblastoma multiforme based on co-expression network analysis.FEBS Open Bio. 2021 Mar;11(3):833-850. doi: 10.1002/2211-5463.13078. Epub 2021 Feb 9. FEBS Open Bio. 2021. PMID: 33423377 Free PMC article.
-
Omeprazole Inhibits Glioblastoma Cell Invasion and Tumor Growth.Cancers (Basel). 2020 Jul 28;12(8):2097. doi: 10.3390/cancers12082097. Cancers (Basel). 2020. PMID: 32731514 Free PMC article.
-
Identification of Candidate Biomarkers and Analysis of Prognostic Values in Oral Squamous Cell Carcinoma.Front Oncol. 2019 Oct 18;9:1054. doi: 10.3389/fonc.2019.01054. eCollection 2019. Front Oncol. 2019. PMID: 31681590 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

