A diagnostic HIV-1 tropism system based on sequence relatedness
- PMID: 25502529
- PMCID: PMC4298515
- DOI: 10.1128/JCM.02762-14
A diagnostic HIV-1 tropism system based on sequence relatedness
Abstract
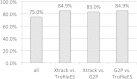
Key clinical studies for HIV coreceptor antagonists have used the phenotyping-based Trofile test. Meanwhile various simpler-to-do genotypic tests have become available that are compatible with standard laboratory equipment and Web-based interpretation tools. However, these systems typically analyze only the most prominent virus sequence in a specimen. We present a new diagnostic HIV tropism test not needing DNA sequencing. The system, XTrack, uses physical properties of DNA duplexes after hybridization of single-stranded HIV-1 env V3 loop probes to the clinical specimen. Resulting "heteroduplexes" possess unique properties driven by sequence relatedness to the reference and resulting in a discrete electrophoretic mobility. A detailed optimization process identified diagnostic probe candidates relating best to a large number of HIV-1 sequences with known tropism. From over 500 V3 sequences representing all main HIV-1 subtypes (Los Alamos database), we obtained a small set of probes to determine the tropism in clinical samples. We found a high concordance with the commercial TrofileES test (84.9%) and the Web-based tool Geno2Pheno (83.0%). Moreover, the new system reveals mixed virus populations, and it was successful on specimens with low virus loads or on provirus from leukocytes. A replicative phenotyping system was used for validation. Our data show that the XTrack test is favorably suitable for routine diagnostics. It detects and dissects mixed virus populations and viral minorities; samples with viral loads (VL) of <200 copies/ml are successfully analyzed. We further expect that the principles of the platform can be adapted also to other sequence-divergent pathogens, such as hepatitis B and C viruses.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures


References
-
- Brumme ZL, Goodrich J, Mayer HB, Brumme CJ, Henrick BM, Wynhoven B, Asselin JJ, Cheung PK, Hogg RS, Montaner JS, Harrigan PR. 2005. Molecular and clinical epidemiology of CXCR4-using HIV-1 in a large population of antiretroviral-naive individuals. J Infect Dis 192:466–474. doi:10.1086/431519. - DOI - PubMed
-
- Huang W, Eshleman SH, Toma J, Fransen S, Stawiski E, Paxinos EE, Whitcomb JM, Young AM, Donnell D, Mmiro F, Musoke P, Guay LA, Jackson JB, Parkin NT, Petropoulos CJ. 2007. Coreceptor tropism in human immunodeficiency virus type 1 subtype D: high prevalence of CXCR4 tropism and heterogeneous composition of viral populations. J Virol 81:7885–7893. doi:10.1128/JVI.00218-07. - DOI - PMC - PubMed
-
- Weiser B, Philpott S, Klimkait T, Burger H, Kitchen C, Burgisser P, Gorgievski M, Perrin L, Piffaretti JC, Ledergerber B. 2008. HIV-1 coreceptor usage and CXCR4-specific viral load predict clinical disease progression during combination antiretroviral therapy. AIDS 22:469–479. doi:10.1097/QAD.0b013e3282f4196c. - DOI - PubMed
-
- Esbjornsson J, Mansson F, Martinez-Arias W, Vincic E, Biague AJ, da Silva ZJ, Fenyo EM, Norrgren H, Medstrand P. 2010. Frequent CXCR4 tropism of HIV-1 subtype A and CRF02_AG during late-stage disease—indication of an evolving epidemic in West Africa. Retrovirology 7:23. doi:10.1186/1742-4690-7-23. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

