Unexpected DNA loss mediated by the DNA binding activity of ribonuclease A
- PMID: 25502562
- PMCID: PMC4263722
- DOI: 10.1371/journal.pone.0115008
Unexpected DNA loss mediated by the DNA binding activity of ribonuclease A
Abstract
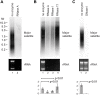
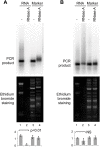
Ribonuclease A (RNase A) is widely used in molecular biology research both for analytical assays and for nucleic acid preparation. The catalytic mechanism of RNase A is well understood and absolutely precludes activity on DNA; however anecdotal reports of DNA degradation by RNase A are not uncommon. Here we describe a mechanism by which RNase A treatment can lead to apparent DNA degradation. This results from the surprising finding that RNase A remains functional in a phenol:chloroform mixture, to our knowledge the only enzyme that survives this highly denaturing solvent environment. Although RNase A does not cleave the DNA backbone it is capable of binding to DNA, forming stable RNase A-DNA complexes that partition to the interphase or organic phase during phenol:chloroform purification. The unexpected survival of the RNase A DNA-binding activity in phenol means that these complexes are not dissolved and a substantial amount of RNase A-bound DNA is permanently removed from the aqueous phase and lost on phase separation. This effect will impact DNA recovery from multiple procedures and is likely to represent a source of sequence bias in genome-wide studies. Our results also indicate that the results of analytical studies performed using RNase A must be considered with care.
Conflict of interest statement
Figures


Similar articles
-
Overexpression, biophysical characterization, and crystallization of ribonuclease I from Escherichia coli, a broad-specificity enzyme in the RNase T2 family.Arch Biochem Biophys. 2001 Jun 1;390(1):42-50. doi: 10.1006/abbi.2001.2359. Arch Biochem Biophys. 2001. PMID: 11368513
-
Degradation of double-stranded RNA by human pancreatic ribonuclease: crucial role of noncatalytic basic amino acid residues.Biochemistry. 2003 Sep 2;42(34):10182-90. doi: 10.1021/bi030040q. Biochemistry. 2003. PMID: 12939146
-
Escherichia coli RNase M is a multiply altered form of RNase I.RNA. 2001 Dec;7(12):1702-7. RNA. 2001. PMID: 11780627 Free PMC article.
-
RNase P from bacteria. Substrate recognition and function of the protein subunit.Mol Biol Rep. 1995-1996;22(2-3):99-109. doi: 10.1007/BF00988713. Mol Biol Rep. 1995. PMID: 8901495 Review.
-
Structural and functional importance of local and global conformational fluctuations in the RNase A superfamily.FEBS J. 2013 Nov;280(22):5596-607. doi: 10.1111/febs.12371. Epub 2013 Jun 24. FEBS J. 2013. PMID: 23763751 Review.
Cited by
-
Genome-wide analysis of DNA replication and DNA double-strand breaks using TrAEL-seq.PLoS Biol. 2021 Mar 24;19(3):e3000886. doi: 10.1371/journal.pbio.3000886. eCollection 2021 Mar. PLoS Biol. 2021. PMID: 33760805 Free PMC article.
-
Chicken muscle mitochondrial content appears co-ordinately regulated and is associated with performance phenotypes.Biol Open. 2017 Jan 15;6(1):50-58. doi: 10.1242/bio.022772. Biol Open. 2017. PMID: 27934661 Free PMC article.
-
Detection of Telomeric DNA:RNA Hybrids Using TeloDRIP-qPCR.Int J Mol Sci. 2020 Dec 21;21(24):9774. doi: 10.3390/ijms21249774. Int J Mol Sci. 2020. PMID: 33371452 Free PMC article.
-
An Optimized Method for the Construction of a DNA Methylome from Small Quantities of Tissue or Purified DNA from Arabidopsis Embryo.Mol Cells. 2021 Aug 31;44(8):602-612. doi: 10.14348/molcells.2021.0084. Mol Cells. 2021. PMID: 34462399 Free PMC article.
-
RNA-DNA hybrid (R-loop) immunoprecipitation mapping: an analytical workflow to evaluate inherent biases.Genome Res. 2017 Jun;27(6):1063-1073. doi: 10.1101/gr.219394.116. Epub 2017 Mar 24. Genome Res. 2017. PMID: 28341774 Free PMC article.
References
-
- Moore S, Stein WH (1973) Chemical structures of pancreatic ribonuclease and deoxyribonuclease. Science 180:458–464. - PubMed
-
- Richards FM (1972) The 1972 nobel prize for chemistry. Science 178:492–493. - PubMed
-
- Anfinsen CB (1973) Principles that govern the folding of protein chains. Science 181:223–230. - PubMed
-
- Hirschmann R, Nutt RF, Veber DF, Vitali RA, Varga SL, et al. (1969) Studies on the total synthesis of an enzyme. V. The preparation of enzymatically active material. J Am Chem Soc 91:507–508. - PubMed
-
- Gutte B, Merrifield RB (1969) The total synthesis of an enzyme with ribonuclease A activity. J Am Chem Soc 91:501–502. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

