Identification of genomic alterations in pancreatic cancer using array-based comparative genomic hybridization
- PMID: 25502777
- PMCID: PMC4263743
- DOI: 10.1371/journal.pone.0114616
Identification of genomic alterations in pancreatic cancer using array-based comparative genomic hybridization
Abstract
Background: Genomic aberration is a common feature of human cancers and also is one of the basic mechanisms that lead to overexpression of oncogenes and underexpression of tumor suppressor genes. Our study aims to identify frequent genomic changes in pancreatic cancer.
Materials and methods: We used array comparative genomic hybridization (array CGH) to identify recurrent genomic alterations and validated the protein expression of selected genes by immunohistochemistry.
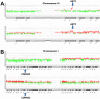
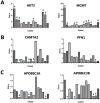
Results: Sixteen gains and thirty-two losses occurred in more than 30% and 60% of the tumors, respectively. High-level amplifications at 7q21.3-q22.1 and 19q13.2 and homozygous deletions at 1p33-p32.3, 1p22.1, 1q22, 3q27.2, 6p22.3, 6p21.31, 12q13.2, 17p13.2, 17q21.31 and 22q13.1 were identified. Especially, amplification of AKT2 was detected in two carcinomas and homozygous deletion of CDKN2C in other two cases. In 15 independent validation samples, we found that AKT2 (19q13.2) and MCM7 (7q22.1) were amplified in 6 and 9 cases, and CAMTA2 (17p13.2) and PFN1 (17p13.2) were homozygously deleted in 3 and 1 cases. AKT2 and MCM7 were overexpressed, and CAMTA2 and PFN1 were underexpressed in pancreatic cancer tissues than in morphologically normal operative margin tissues. Both GISTIC and Genomic Workbench software identified 22q13.1 containing APOBEC3A and APOBEC3B as the only homozygous deletion region. And the expression levels of APOBEC3A and APOBEC3B were significantly lower in tumor tissues than in morphologically normal operative margin tissues. Further validation showed that overexpression of PSCA was significantly associated with lymph node metastasis, and overexpression of HMGA2 was significantly associated with invasive depth of pancreatic cancer.
Conclusion: These recurrent genomic changes may be useful for revealing the mechanism of pancreatic carcinogenesis and providing candidate biomarkers.
Conflict of interest statement
Figures




Similar articles
-
Genomic landscape of pancreatic neuroendocrine tumors.World J Gastroenterol. 2014 Dec 14;20(46):17498-506. doi: 10.3748/wjg.v20.i46.17498. World J Gastroenterol. 2014. PMID: 25516664 Free PMC article.
-
Lacrimal gland pleomorphic adenoma and carcinoma ex pleomorphic adenoma: genomic profiles, gene fusions, and clinical characteristics.Ophthalmology. 2014 May;121(5):1125-33. doi: 10.1016/j.ophtha.2013.11.037. Epub 2014 Jan 24. Ophthalmology. 2014. PMID: 24468654
-
Array-based comparative genomic hybridization identifies localized DNA amplifications and homozygous deletions in pancreatic cancer.Neoplasia. 2005 Jun;7(6):556-62. doi: 10.1593/neo.04586. Neoplasia. 2005. PMID: 16036106 Free PMC article.
-
Genome-wide analysis of pancreatic cancer using microarray-based techniques.Pancreatology. 2009;9(1-2):13-24. doi: 10.1159/000178871. Epub 2008 Dec 12. Pancreatology. 2009. PMID: 19077451 Review.
-
Novel technology for detection of genomic and transcriptional alterations in pancreatic cancer.Ann Oncol. 1999;10 Suppl 4:64-8. Ann Oncol. 1999. PMID: 10436788 Review.
Cited by
-
Role of the single deaminase domain APOBEC3A in virus restriction, retrotransposition, DNA damage and cancer.J Gen Virol. 2016 Jan;97(1):1-17. doi: 10.1099/jgv.0.000320. Epub 2015 Oct 20. J Gen Virol. 2016. PMID: 26489798 Free PMC article. Review.
-
Transcriptomic Analysis of Laser Capture Microdissected Tumors Reveals Cancer- and Stromal-Specific Molecular Subtypes of Pancreatic Ductal Adenocarcinoma.Clin Cancer Res. 2021 Apr 15;27(8):2314-2325. doi: 10.1158/1078-0432.CCR-20-1039. Epub 2021 Feb 5. Clin Cancer Res. 2021. PMID: 33547202 Free PMC article.
-
Phosphoinositide 3-Kinase Signaling Pathway in Pancreatic Ductal Adenocarcinoma Progression, Pathogenesis, and Therapeutics.Front Physiol. 2018 Apr 4;9:335. doi: 10.3389/fphys.2018.00335. eCollection 2018. Front Physiol. 2018. PMID: 29670543 Free PMC article. Review.
-
Weighted gene co-expression network analysis reveals key genes involved in pancreatic ductal adenocarcinoma development.Cell Oncol (Dordr). 2016 Aug;39(4):379-88. doi: 10.1007/s13402-016-0283-7. Epub 2016 May 30. Cell Oncol (Dordr). 2016. PMID: 27240826
-
APOBEC: A molecular driver in cervical cancer pathogenesis.Cancer Lett. 2021 Jan 1;496:104-116. doi: 10.1016/j.canlet.2020.10.004. Epub 2020 Oct 7. Cancer Lett. 2021. PMID: 33038491 Free PMC article. Review.
References
-
- Jemal A, Siegel R, Xu J, Ward E (2010) Cancer statistics, 2010. CA Cancer J Clin 60:277–300. - PubMed
-
- Negrini S, Gorgoulis VG, Halazonetis TD (2010) Genomic instability–an evolving hallmark of cancer. Nat Rev Mol Cell Biol 11:220–228. - PubMed
-
- Harada T, Okita K, Shiraishi K, Kusano N, Kondoh S, et al. (2002) Interglandular cytogenetic heterogeneity detected by comparative genomic hybridization in pancreatic cancer. Cancer Res 62:835–839. - PubMed
-
- Kitoh H, Ryozawa S, Harada T, Kondoh S, Furuya T, et al. (2005) Comparative genomic hybridization analysis for pancreatic cancer specimens obtained by endoscopic ultrasonography-guided fine-needle aspiration. J Gastroenterol 40:511–517. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

