Different binding motifs of the celiac disease-associated HLA molecules DQ2.5, DQ2.2, and DQ7.5 revealed by relative quantitative proteomics of endogenous peptide repertoires
- PMID: 25502872
- PMCID: PMC4297300
- DOI: 10.1007/s00251-014-0819-9
Different binding motifs of the celiac disease-associated HLA molecules DQ2.5, DQ2.2, and DQ7.5 revealed by relative quantitative proteomics of endogenous peptide repertoires
Abstract
Celiac disease is caused by intolerance to cereal gluten proteins, and HLA-DQ molecules are involved in the disease pathogenesis by presentation of gluten peptides to CD4(+) T cells. The α- or β-chain sharing HLA molecules DQ2.5, DQ2.2, and DQ7.5 display different risks for the disease. It was recently demonstrated that T cells of DQ2.5 and DQ2.2 patients recognize distinct sets of gluten epitopes, suggesting that these two DQ2 variants select different peptides for display. To explore whether this is the case, we performed a comprehensive comparison of the endogenous self-peptides bound to HLA-DQ molecules of B-lymphoblastoid cell lines. Peptides were eluted from affinity-purified HLA molecules of nine cell lines and subjected to quadrupole orbitrap mass spectrometry and MaxQuant software analysis. Altogether, 12,712 endogenous peptides were identified at very different relative abundances. Hierarchical clustering of normalized quantitative data demonstrated significant differences in repertoires of peptides between the three DQ variant molecules. The neural network-based method, NNAlign, was used to identify peptide-binding motifs. The binding motifs of DQ2.5 and DQ7.5 concurred with previously established binding motifs. The binding motif of DQ2.2 was strikingly different from that of DQ2.5 with position P3 being a major anchor having a preference for threonine and serine. This is notable as three recently identified epitopes of gluten recognized by T cells of DQ2.2 celiac patients harbor serine at position P3. This study demonstrates that relative quantitative comparison of endogenous peptides sampled from our protein metabolism by HLA molecules provides clues to understand HLA association with disease.
Figures

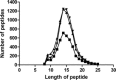

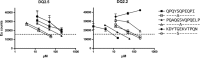
References
-
- Efron B. Size, power and false discovery rates. Ann Stat. 2007;35:1351–1377. doi: 10.1214/009053606000001460. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

