Genome-wide detection of DNA double-stranded breaks induced by engineered nucleases
- PMID: 25503383
- PMCID: PMC4320661
- DOI: 10.1038/nbt.3101
Genome-wide detection of DNA double-stranded breaks induced by engineered nucleases
Abstract
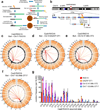
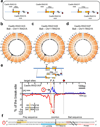
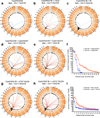
Although great progress has been made in the characterization of the off-target effects of engineered nucleases, sensitive and unbiased genome-wide methods for the detection of off-target cleavage events and potential collateral damage are still lacking. Here we describe a linear amplification-mediated modification of a previously published high-throughput, genome-wide, translocation sequencing (HTGTS) method that robustly detects DNA double-stranded breaks (DSBs) generated by engineered nucleases across the human genome based on their translocation to other endogenous or ectopic DSBs. HTGTS with different Cas9:sgRNA or TALEN nucleases revealed off-target hotspot numbers for given nucleases that ranged from a few or none to dozens or more, and extended the number of known off-targets for certain previously characterized nucleases more than tenfold. We also identified translocations between bona fide nuclease targets on homologous chromosomes, an undesired collateral effect that has not been described previously. Finally, HTGTS confirmed that the Cas9D10A paired nickase approach suppresses off-target cleavage genome-wide.
Figures


Comment in
-
Mapping the precision of genome editing.Nat Biotechnol. 2015 Feb;33(2):150-2. doi: 10.1038/nbt.3142. Nat Biotechnol. 2015. PMID: 25658281 No abstract available.
-
Collateral DNA damage produced by genome-editing drones: exception or rule?Mol Cell. 2015 May 21;58(4):565-7. doi: 10.1016/j.molcel.2015.05.012. Mol Cell. 2015. PMID: 26000841 Free PMC article.
References
-
- Kim H, Kim JS. A guide to genome engineering with programmable nucleases. Nat. Rev. Genet. 2014;15:321–334. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

