UPF2, a nonsense-mediated mRNA decay factor, is required for prepubertal Sertoli cell development and male fertility by ensuring fidelity of the transcriptome
- PMID: 25503407
- PMCID: PMC4302838
- DOI: 10.1242/dev.115642
UPF2, a nonsense-mediated mRNA decay factor, is required for prepubertal Sertoli cell development and male fertility by ensuring fidelity of the transcriptome
Abstract
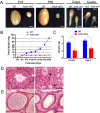
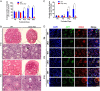
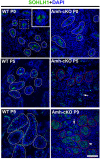
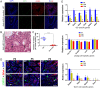
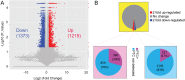
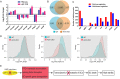
Nonsense-mediated mRNA decay (NMD) represents a highly conserved RNA surveillance mechanism through which mRNA transcripts bearing premature termination codons (PTCs) are selectively degraded to maintain transcriptomic fidelity in the cell. Numerous in vitro studies have demonstrated the importance of the NMD pathway; however, evidence supporting its physiological necessity has only just started to emerge. Here, we report that ablation of Upf2, which encodes a core NMD factor, in murine embryonic Sertoli cells (SCs) leads to severe testicular atrophy and male sterility owing to rapid depletion of both SCs and germ cells during prepubertal testicular development. RNA-Seq and bioinformatic analyses revealed impaired transcriptomic homeostasis in SC-specific Upf2 knockout testes, characterized by an accumulation of PTC-containing transcripts and the transcriptome-wide dysregulation of genes encoding splicing factors and key proteins essential for SC fate control. Our data demonstrate an essential role of UPF2-mediated NMD in prepubertal SC development and male fertility.
Keywords: 3′UTR shortening; Alternative splicing; Gonocyte; Mouse; Nonsense-mediated mRNA decay; Premature termination codon; RNA-Seq; Sertoli cell; Spermatogenesis; Sterility; Testis.
© 2015. Published by The Company of Biologists Ltd.
Figures

Similar articles
-
UPF2-Dependent Nonsense-Mediated mRNA Decay Pathway Is Essential for Spermatogenesis by Selectively Eliminating Longer 3'UTR Transcripts.PLoS Genet. 2016 May 5;12(5):e1005863. doi: 10.1371/journal.pgen.1005863. eCollection 2016 May. PLoS Genet. 2016. PMID: 27149259 Free PMC article.
-
Nonsense in the testis: multiple roles for nonsense-mediated decay revealed in male reproduction.Biol Reprod. 2017 May 1;96(5):939-947. doi: 10.1093/biolre/iox033. Biol Reprod. 2017. PMID: 28444146 Free PMC article. Review.
-
Upf proteins: highly conserved factors involved in nonsense mRNA mediated decay.Mol Biol Rep. 2018 Feb;45(1):39-55. doi: 10.1007/s11033-017-4139-7. Epub 2017 Dec 27. Mol Biol Rep. 2018. PMID: 29282598 Review.
-
Contribution of copy number variants involving nonsense-mediated mRNA decay pathway genes to neuro-developmental disorders.Hum Mol Genet. 2013 May 1;22(9):1816-25. doi: 10.1093/hmg/ddt035. Epub 2013 Jan 31. Hum Mol Genet. 2013. PMID: 23376982
-
hnRNPU in Sertoli cells cooperates with WT1 and is essential for testicular development by modulating transcriptional factors Sox8/9.Theranostics. 2021 Oct 25;11(20):10030-10046. doi: 10.7150/thno.66819. eCollection 2021. Theranostics. 2021. PMID: 34815802 Free PMC article.
Cited by
-
Characterization of rodent Sertoli cell primary cultures.Mol Reprod Dev. 2020 Aug;87(8):857-870. doi: 10.1002/mrd.23402. Epub 2020 Aug 2. Mol Reprod Dev. 2020. PMID: 32743879 Free PMC article. Review.
-
Characterization of a rhabdomyosarcoma reveals a critical role for SMG7 in cancer cell viability and tumor growth.Sci Rep. 2023 Jun 22;13(1):10152. doi: 10.1038/s41598-023-36568-5. Sci Rep. 2023. PMID: 37349371 Free PMC article.
-
MicroRNAs control mRNA fate by compartmentalization based on 3' UTR length in male germ cells.Genome Biol. 2017 Jun 15;18(1):105. doi: 10.1186/s13059-017-1243-x. Genome Biol. 2017. PMID: 28615029 Free PMC article.
-
Physiological Consequences of Nonsense-Mediated Decay and Its Role in Adaptive Responses.Biomedicines. 2024 May 16;12(5):1110. doi: 10.3390/biomedicines12051110. Biomedicines. 2024. PMID: 38791071 Free PMC article. Review.
-
Nonsense-Mediated mRNA Decay: Mechanistic Insights and Physiological Significance.Mol Biotechnol. 2024 Nov;66(11):3077-3091. doi: 10.1007/s12033-023-00927-4. Epub 2023 Nov 6. Mol Biotechnol. 2024. PMID: 37930508 Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

