Pre-treatment whole blood gene expression is associated with 14-week response assessed by dynamic contrast enhanced magnetic resonance imaging in infliximab-treated rheumatoid arthritis patients
- PMID: 25504080
- PMCID: PMC4264695
- DOI: 10.1371/journal.pone.0113937
Pre-treatment whole blood gene expression is associated with 14-week response assessed by dynamic contrast enhanced magnetic resonance imaging in infliximab-treated rheumatoid arthritis patients
Abstract
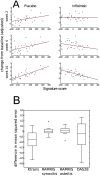
Approximately 30% of rheumatoid arthritis patients achieve inadequate response to anti-TNF biologics. Attempts to identify molecular biomarkers predicting response have met with mixed success. This may be attributable, in part, to the variable and subjective disease assessment endpoints with large placebo effects typically used to classify patient response. Sixty-one patients with active RA despite methotrexate treatment, and with MRI-documented synovitis, were randomized to receive infliximab or placebo. Blood was collected at baseline and genome-wide transcription in whole blood was measured using microarrays. The primary endpoint in this study was determined by measuring the transfer rate constant (Ktrans) of a gadolinium-based contrast agent from plasma to synovium using MRI. Secondary endpoints included repeated clinical assessments with DAS28(CRP), and assessments of osteitis and synovitis by the RAMRIS method. Infliximab showed greater decrease from baseline in DCE-MRI Ktrans of wrist and MCP at all visits compared with placebo (P<0.001). Statistical analysis was performed to identify genes associated with treatment-specific 14-week change in Ktrans. The 256 genes identified were used to derive a gene signature score by averaging their log expression within each patient. The resulting score correlated with improvement of Ktrans in infliximab-treated patients and with deterioration of Ktrans in placebo-treated subjects. Poor responders showed high expression of activated B-cell genes whereas good responders exhibited a gene expression pattern consistent with mobilization of neutrophils and monocytes and high levels of reticulated platelets. This gene signature was significantly associated with clinical response in two previously published whole blood gene expression studies using anti-TNF therapies. These data provide support for the hypothesis that anti-TNF inadequate responders comprise a distinct molecular subtype of RA characterized by differences in pre-treatment blood mRNA expression. They also highlight the importance of placebo controls and robust, objective endpoints in biomarker discovery.
Trial registration: ClinicalTrials.gov NCT01313520.
Conflict of interest statement
Figures

References
-
- Hyrich KL, Watson KD, Silman AJ, Symmons DPM (2006) Predictors of response to anti-TNF-alpha therapy among patients with rheumatoid arthritis: results from the British Society for Rheumatology Biologics Register. Rheumatology (Oxford) 45:1558–1565. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

