Genetic and phenotypic intra-species variation in Candida albicans
- PMID: 25504520
- PMCID: PMC4352881
- DOI: 10.1101/gr.174623.114
Genetic and phenotypic intra-species variation in Candida albicans
Abstract
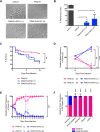
Candida albicans is a commensal fungus of the human gastrointestinal tract and a prevalent opportunistic pathogen. To examine diversity within this species, extensive genomic and phenotypic analyses were performed on 21 clinical C. albicans isolates. Genomic variation was evident in the form of polymorphisms, copy number variations, chromosomal inversions, subtelomeric hypervariation, loss of heterozygosity (LOH), and whole or partial chromosome aneuploidies. All 21 strains were diploid, although karyotypic changes were present in eight of the 21 isolates, with multiple strains being trisomic for Chromosome 4 or Chromosome 7. Aneuploid strains exhibited a general fitness defect relative to euploid strains when grown under replete conditions. All strains were also heterozygous, yet multiple, distinct LOH tracts were present in each isolate. Higher overall levels of genome heterozygosity correlated with faster growth rates, consistent with increased overall fitness. Genes with the highest rates of amino acid substitutions included many cell wall proteins, implicating fast evolving changes in cell adhesion and host interactions. One clinical isolate, P94015, presented several striking properties including a novel cellular phenotype, an inability to filament, drug resistance, and decreased virulence. Several of these properties were shown to be due to a homozygous nonsense mutation in the EFG1 gene. Furthermore, loss of EFG1 function resulted in increased fitness of P94015 in a commensal model of infection. Our analysis therefore reveals intra-species genetic and phenotypic differences in C. albicans and delineates a natural mutation that alters the balance between commensalism and pathogenicity.
© 2015 Hirakawa et al.; Published by Cold Spring Harbor Laboratory Press.
Figures





References
Publication types
MeSH terms
Associated data
- BioProject/PRJNA193498
- dbSNP/1456786277
- dbSNP/1456786278
- dbSNP/1456786279
- dbSNP/1456786280
- dbSNP/1456786281
- dbSNP/1456786282
- dbSNP/1456786283
- dbSNP/1456786284
- dbSNP/1456786285
- dbSNP/1456786286
- dbSNP/1456786287
- dbSNP/1456786288
- dbSNP/1456786289
- dbSNP/1456786290
- dbSNP/1456786291
- dbSNP/1456786292
- dbSNP/1456786293
- dbSNP/1456786294
- dbSNP/1456786295
- dbSNP/1456786296
- dbSNP/1456786297
- dbSNP/1456786298
- dbSNP/1456786299
- dbSNP/1456786300
- dbSNP/1456786301
- dbSNP/1456786302
- dbSNP/1456786303
- dbSNP/1456786304
- dbSNP/1456786305
- dbSNP/1456786306
- dbSNP/1456786307
- dbSNP/1456786308
- dbSNP/1456786309
- dbSNP/1456786310
- dbSNP/1456786311
- dbSNP/1456786312
- dbSNP/1456786313
- dbSNP/1456786314
- dbSNP/1456786315
- dbSNP/1456786316
- dbSNP/1456786317
- dbSNP/1456786318
- dbSNP/1456786319
- dbSNP/1456786320
- dbSNP/1456786321
- dbSNP/1456786322
- dbSNP/1456786323
- dbSNP/1456786324
- dbSNP/1456786325
- dbSNP/1456786326
- dbSNP/1456786327
- dbSNP/1456786328
- dbSNP/1456786329
- dbSNP/1456786330
- dbSNP/1456786331
- dbSNP/1456786332
- dbSNP/1456786333
- dbSNP/1456786334
- dbSNP/1456786335
- dbSNP/1456786336
- dbSNP/1456786337
- dbSNP/1456786338
- dbSNP/1456786339
- dbSNP/1456786340
- dbSNP/1456786341
- dbSNP/1456786342
- dbSNP/1456786343
- dbSNP/1456786344
- dbSNP/1456786345
- dbSNP/1456786346
- dbSNP/1456786347
- dbSNP/1456786348
- dbSNP/1456786349
- dbSNP/1456786350
- dbSNP/1456786351
- dbSNP/1456786352
- dbSNP/1456786353
- dbSNP/1456786354
- dbSNP/1456786355
- dbSNP/1456786356
- dbSNP/1456786357
- dbSNP/1456786358
- dbSNP/1456786359
- dbSNP/1456786360
- dbSNP/1456786361
- dbSNP/1456786362
- dbSNP/1456786363
- dbSNP/1456786364
- dbSNP/1456786365
- dbSNP/1456786366
- dbSNP/1456786367
- dbSNP/1456786368
- dbSNP/1456786369
- dbSNP/1456786370
- dbSNP/1456786371
- dbSNP/1456786372
- dbSNP/1456786373
- dbSNP/1456786374
- dbSNP/1456786375
- dbSNP/1456786376
- dbSNP/1456786377
- dbSNP/1456786378
- dbSNP/1456786379
- dbSNP/1456786380
- dbSNP/1456786381
- dbSNP/1456786382
- dbSNP/1456786383
- dbSNP/1456786384
- dbSNP/1456786385
- dbSNP/1456786386
- dbSNP/1456786387
- dbSNP/1456786388
- dbSNP/1456786389
- dbSNP/1456786390
- dbSNP/1456786391
- dbSNP/1456786392
- dbSNP/1456786393
- dbSNP/1456786394
- dbSNP/1456786395
- dbSNP/1456786396
- dbSNP/1456786397
- dbSNP/1456786398
- dbSNP/1456786399
- dbSNP/1456786400
- dbSNP/1456786401
- dbSNP/1456786402
- dbSNP/1456786403
- dbSNP/1456786404
- dbSNP/1456786405
- dbSNP/1456786406
- dbSNP/1456786407
- dbSNP/1456786408
- dbSNP/1456786409
- dbSNP/1456786410
- dbSNP/1456786411
- dbSNP/1456786412
- dbSNP/1456786413
- dbSNP/1456786414
- dbSNP/1456786415
- dbSNP/1456786416
- dbSNP/1456786417
- dbSNP/1456786418
- dbSNP/1456786419
- dbSNP/1456786420
- dbSNP/1456786421
- dbSNP/1456786422
- dbSNP/1456786423
- dbSNP/1456786424
- dbSNP/1456786425
- dbSNP/1456786426
- dbSNP/1456786427
- dbSNP/1456786428
- dbSNP/1456786429
- dbSNP/1456786430
- dbSNP/1456786431
- dbSNP/1456786432
- dbSNP/1456786433
- dbSNP/1456786434
- dbSNP/1456786435
- dbSNP/1456786436
- dbSNP/1456786437
- dbSNP/1456786438
- dbSNP/1456786439
- dbSNP/1456786440
- dbSNP/1456786441
- dbSNP/1456786442
- dbSNP/1456786443
- dbSNP/1456786444
- dbSNP/1456786445
- dbSNP/1456786446
- dbSNP/1456786447
- dbSNP/1456786448
- dbSNP/1456786449
- dbSNP/1456786450
- dbSNP/1456786451
- dbSNP/1456786452
- dbSNP/1456786453
- dbSNP/1456786454
- dbSNP/1456786455
- dbSNP/1456786456
- dbSNP/1456786457
- dbSNP/1456786458
- dbSNP/1456786459
- dbSNP/1456786460
- dbSNP/1456786461
- dbSNP/1456786462
- dbSNP/1456786463
- dbSNP/1456786464
- dbSNP/1456786465
- dbSNP/1456786466
- dbSNP/1456786467
- dbSNP/1456786468
- dbSNP/1456786469
- dbSNP/1456786470
- dbSNP/1456786471
- dbSNP/1456786472
- dbSNP/1456786473
- dbSNP/1456786474
- dbSNP/1456786475
- dbSNP/1456786476
- dbSNP/1456786477
- dbSNP/1456786478
- dbSNP/1456786479
- dbSNP/1456786480
- dbSNP/1456786481
- dbSNP/1456786482
- dbSNP/1456786483
- dbSNP/1456786484
- dbSNP/1456786485
- dbSNP/1456786486
- dbSNP/1456786487
- dbSNP/1456786488
- dbSNP/1456786489
- dbSNP/1456786490
- dbSNP/1456786491
- dbSNP/1456786492
- dbSNP/1456786493
- dbSNP/1456786494
- dbSNP/1456786495
- dbSNP/1456786496
- dbSNP/1456786497
- dbSNP/1456786498
- dbSNP/1456786499
- dbSNP/1456786500
- dbSNP/1456786501
- dbSNP/1456786502
- dbSNP/1456786503
- dbSNP/1456786504
- dbSNP/1456786505
- dbSNP/1456786506
- dbSNP/1456786507
- dbSNP/1456786508
- dbSNP/1456786509
- dbSNP/1456786510
- dbSNP/1456786511
- dbSNP/1456786512
- dbSNP/1456786513
- dbSNP/1456786514
- dbSNP/1456786515
- dbSNP/1456786516
- dbSNP/1456786517
- dbSNP/1456786518
- dbSNP/1456786519
- dbSNP/1456786520
- dbSNP/1456786521
- dbSNP/1456786522
- dbSNP/1456786523
- dbSNP/1456786524
- dbSNP/1456786525
- dbSNP/1456786526
- dbSNP/1456786527
- dbSNP/1456786528
- dbSNP/1456786529
- dbSNP/1456786530
- dbSNP/1456786531
- dbSNP/1456786532
- dbSNP/1456786533
- dbSNP/1456786534
- dbSNP/1456786535
- dbSNP/1456786536
- dbSNP/1456786537
- dbSNP/1456786538
- dbSNP/1456786539
- dbSNP/1456786540
- dbSNP/1456786541
- dbSNP/1456786542
- dbSNP/1456786543
- dbSNP/1456786544
- dbSNP/1456786545
- dbSNP/1456786546
- dbSNP/1456786547
- dbSNP/1456786548
- dbSNP/1456786549
- dbSNP/1456786550
- dbSNP/1456786551
- dbSNP/1456786552
- dbSNP/1456786553
- dbSNP/1456786554
- dbSNP/1456786555
- dbSNP/1456786556
- dbSNP/1456786557
- dbSNP/1456786558
- dbSNP/1456786559
- dbSNP/1456786560
- dbSNP/1456786561
- dbSNP/1456786562
- dbSNP/1456786563
- dbSNP/1456786564
- dbSNP/1456786565
- dbSNP/1456786566
- dbSNP/1456786567
- dbSNP/1456786568
- dbSNP/1456786569
- dbSNP/1456786570
- dbSNP/1456786571
- dbSNP/1456786572
- dbSNP/1456786573
- dbSNP/1456786574
- dbSNP/1456786575
- dbSNP/1456786576
- dbSNP/1456786577
- dbSNP/1456786578
- dbSNP/1456786579
- dbSNP/1456786580
- dbSNP/1456786581
- dbSNP/1456786582
- dbSNP/1456786583
- dbSNP/1456786584
- dbSNP/1456786585
- dbSNP/1456786586
- dbSNP/1456786587
- dbSNP/1456786588
- dbSNP/1456786589
- dbSNP/1456786590
- dbSNP/1456786591
- dbSNP/1456786592
- dbSNP/1456786593
- dbSNP/1456786594
- dbSNP/1456786595
- dbSNP/1456786596
- dbSNP/1456786597
- dbSNP/1456786598
- dbSNP/1456786599
- dbSNP/1456786600
- dbSNP/1456786601
- dbSNP/1456786602
- dbSNP/1456786603
- dbSNP/1456786604
- dbSNP/1456786605
- dbSNP/1456786606
- dbSNP/1456786607
- dbSNP/1456786608
- dbSNP/1456786609
- dbSNP/1456786610
- dbSNP/1456786611
- dbSNP/1456786612
- dbSNP/1456786613
- dbSNP/1456786614
- dbSNP/1456786615
- dbSNP/1456786616
- dbSNP/1456786617
- dbSNP/1456786618
- dbSNP/1456786619
- dbSNP/1456786620
- dbSNP/1456786621
- dbSNP/1456786622
- dbSNP/1456786623
- dbSNP/1456786624
- dbSNP/1456786625
- dbSNP/1456786626
- dbSNP/1456786627
- dbSNP/1456786628
- dbSNP/1456786629
- dbSNP/1456786630
- dbSNP/1456786631
- dbSNP/1456786632
- dbSNP/1456786633
- dbSNP/1456786634
- dbSNP/1456786635
- dbSNP/1456786636
- dbSNP/1456786637
- dbSNP/1456786638
- dbSNP/1456786639
- dbSNP/1456786640
- dbSNP/1456786641
- dbSNP/1456786642
- dbSNP/1456786643
- dbSNP/1456786644
- dbSNP/1456786645
- dbSNP/1456786646
- dbSNP/1456786647
- dbSNP/1456786648
- dbSNP/1456786649
- dbSNP/1456786650
- dbSNP/1456786651
- dbSNP/1456786652
- dbSNP/1456786653
- dbSNP/1456786654
- dbSNP/1456786655
- dbSNP/1456786656
- dbSNP/1456786657
- dbSNP/1456786658
- dbSNP/1456786659
- dbSNP/1456786660
- dbSNP/1456786661
- dbSNP/1456786662
- dbSNP/1456786663
- dbSNP/1456786664
- dbSNP/1456786665
- dbSNP/1456786666
- dbSNP/1456786667
- dbSNP/1456786668
- dbSNP/1456786669
- dbSNP/1456786670
- dbSNP/1456786671
- dbSNP/1456786672
- dbSNP/1456786673
- dbSNP/1456786674
- dbSNP/1456786675
- dbSNP/1456786676
- dbSNP/1456786677
- dbSNP/1456786678
- dbSNP/1456786679
- dbSNP/1456786680
- dbSNP/1456786681
- dbSNP/1456786682
- dbSNP/1456786683
- dbSNP/1456786684
- dbSNP/1456786685
- dbSNP/1456786686
- dbSNP/1456786687
- dbSNP/1456786688
- dbSNP/1456786689
- dbSNP/1456786690
- dbSNP/1456786691
- dbSNP/1456786692
- dbSNP/1456786693
- dbSNP/1456786694
- dbSNP/1456786695
- dbSNP/1456786696
- dbSNP/1456786697
- dbSNP/1456786698
- dbSNP/1456786699
- dbSNP/1456786700
- dbSNP/1456786701
- dbSNP/1456786702
- dbSNP/1456786703
- dbSNP/1456786704
- dbSNP/1456786705
- dbSNP/1456786706
- dbSNP/1456786707
- dbSNP/1456786708
- dbSNP/1456786709
- dbSNP/1456786710
- dbSNP/1456786711
- dbSNP/1456786712
- dbSNP/1456786713
- dbSNP/1456786714
- dbSNP/1456786715
- dbSNP/1456786716
- dbSNP/1456786717
- dbSNP/1456786718
- dbSNP/1456786719
- dbSNP/1456786720
- dbSNP/1456786721
- dbSNP/1456786722
- dbSNP/1456786723
- dbSNP/1456786724
- dbSNP/1456786725
- dbSNP/1456786726
- dbSNP/1456786727
- dbSNP/1456786728
- dbSNP/1456786729
- dbSNP/1456786730
- dbSNP/1456786731
- dbSNP/1456786732
- dbSNP/1456786733
- dbSNP/1456786734
- dbSNP/1456786735
- dbSNP/1456786736
- dbSNP/1456786737
- dbSNP/1456786738
- dbSNP/1456786739
- dbSNP/1456786740
- dbSNP/1456786741
- dbSNP/1456786742
- dbSNP/1456786743
- dbSNP/1456786744
- dbSNP/1456786745
- dbSNP/1456786746
- dbSNP/1456786747
- dbSNP/1456786748
- dbSNP/1456786749
- dbSNP/1456786750
- dbSNP/1456786751
- dbSNP/1456786752
- dbSNP/1456786753
- dbSNP/1456786754
- dbSNP/1456786755
- dbSNP/1456786756
- dbSNP/1456786757
- dbSNP/1456786758
- dbSNP/1456786759
- dbSNP/1456786760
- dbSNP/1456786761
- dbSNP/1456786762
- dbSNP/1456786763
- dbSNP/1456786764
- dbSNP/1456786765
- dbSNP/1456786766
- dbSNP/1456786767
- dbSNP/1456786768
- dbSNP/1456786769
- dbSNP/1456786770
- dbSNP/1456786771
- dbSNP/1456786772
- dbSNP/1456786773
- dbSNP/1456786774
- dbSNP/1456786775
- dbSNP/1456786776
- dbSNP/1456786777
- dbSNP/1456786778
- dbSNP/1456786779
- dbSNP/1456786780
- dbSNP/1456786781
- dbSNP/1456786782
- dbSNP/1456786783
- dbSNP/1456786784
- dbSNP/1456786785
- dbSNP/1456786786
- dbSNP/1456786787
- dbSNP/1456786788
- dbSNP/1456786789
- dbSNP/1456786790
- dbSNP/1456786791
- dbSNP/1456786792
- dbSNP/1456786793
- dbSNP/1456786794
- dbSNP/1456786795
- dbSNP/1456786796
- dbSNP/1456786797
- dbSNP/1456786798
- dbSNP/1456786799
- dbSNP/1456786800
- dbSNP/1456786801
- dbSNP/1456786802
- dbSNP/1456786803
- dbSNP/1456786804
- dbSNP/1456786805
- dbSNP/1456786806
- dbSNP/1456786807
- dbSNP/1456786808
- dbSNP/1456786809
- dbSNP/1456786810
- dbSNP/1456786811
- dbSNP/1456786812
- dbSNP/1456786813
- dbSNP/1456786814
- dbSNP/1456786815
- dbSNP/1456786816
- dbSNP/1456786817
- dbSNP/1456786818
- dbSNP/1456786819
- dbSNP/1456786820
- dbSNP/1456786821
- dbSNP/1456786822
- dbSNP/1456786823
- dbSNP/1456786824
- dbSNP/1456786825
- dbSNP/1456786826
- dbSNP/1456786827
- dbSNP/1456786828
- dbSNP/1456786829
- dbSNP/1456786830
- dbSNP/1456786831
- dbSNP/1456786832
- dbSNP/1456786833
- dbSNP/1456786834
- dbSNP/1456786835
- dbSNP/1456786836
- dbSNP/1456786837
- dbSNP/1456786838
- dbSNP/1456786839
- dbSNP/1456786840
- dbSNP/1456786841
- dbSNP/1456786842
- dbSNP/1456786843
- dbSNP/1456786844
- dbSNP/1456786845
- dbSNP/1456786846
- dbSNP/1456786847
- dbSNP/1456786848
- dbSNP/1456786849
- dbSNP/1456786850
- dbSNP/1456786851
- dbSNP/1456786852
- dbSNP/1456786853
- dbSNP/1456786854
- dbSNP/1456786855
- dbSNP/1456786856
- dbSNP/1456786857
- dbSNP/1456786858
- dbSNP/1456786859
- dbSNP/1456786860
- dbSNP/1456786861
- dbSNP/1456786862
- dbSNP/1456786863
- dbSNP/1456786864
- dbSNP/1456786865
- dbSNP/1456786866
- dbSNP/1456786867
- dbSNP/1456786868
- dbSNP/1456786869
- dbSNP/1456786870
- dbSNP/1456786871
- dbSNP/1456786872
- dbSNP/1456786873
- dbSNP/1456786874
- dbSNP/1456786875
- dbSNP/1456786876
- dbSNP/1456786877
- dbSNP/1456786878
- dbSNP/1456786879
- dbSNP/1456786880
- dbSNP/1456786881
- dbSNP/1456786882
- dbSNP/1456786883
- dbSNP/1456786884
- dbSNP/1456786885
- dbSNP/1456786886
- dbSNP/1456786887
- dbSNP/1456786888
- dbSNP/1456786889
- dbSNP/1456786890
- dbSNP/1456786891
- dbSNP/1456786892
- dbSNP/1456786893
- dbSNP/1456786894
- dbSNP/1456786895
- dbSNP/1456786896
- dbSNP/1456786897
- dbSNP/1456786898
- dbSNP/1456786899
- dbSNP/1456786900
- dbSNP/1456786901
- dbSNP/1456786902
- dbSNP/1456786903
- dbSNP/1456786904
- dbSNP/1456786905
- dbSNP/1456786906
- dbSNP/1456786907
- dbSNP/1456786908
- dbSNP/1456786909
- dbSNP/1456786910
- dbSNP/1456786911
- dbSNP/1456786912
- dbSNP/1456786913
- dbSNP/1456786914
- dbSNP/1456786915
- dbSNP/1456786916
- dbSNP/1456786917
- dbSNP/1456786918
- dbSNP/1456786919
- dbSNP/1456786920
- dbSNP/1456786921
- dbSNP/1456786922
- dbSNP/1456786923
- dbSNP/1456786924
- dbSNP/1456786925
- dbSNP/1456786926
- dbSNP/1456786927
- dbSNP/1456786928
- dbSNP/1456786929
- dbSNP/1456786930
- dbSNP/1456786931
- dbSNP/1456786932
- dbSNP/1456786933
- dbSNP/1456786934
- dbSNP/1456786935
- dbSNP/1456786936
- dbSNP/1456786937
- dbSNP/1456786938
- dbSNP/1456786939
- dbSNP/1456786940
- dbSNP/1456786941
- dbSNP/1456786942
- dbSNP/1456786943
- dbSNP/1456786944
- dbSNP/1456786945
- dbSNP/1456786946
- dbSNP/1456786947
- dbSNP/1456786948
- dbSNP/1456786949
- dbSNP/1456786950
- dbSNP/1456786951
- dbSNP/1456786952
- dbSNP/1456786953
- dbSNP/1456786954
- dbSNP/1456786955
- dbSNP/1456786956
- dbSNP/1456786957
- dbSNP/1456786958
- dbSNP/1456786959
- dbSNP/1456786960
- dbSNP/1456786961
- dbSNP/1456786962
- dbSNP/1456786963
- dbSNP/1456786964
- dbSNP/1456786965
- dbSNP/1456786966
- dbSNP/1456786967
- dbSNP/1456786968
- dbSNP/1456786969
- dbSNP/1456786970
- dbSNP/1456786971
- dbSNP/1456786972
- dbSNP/1456786973
- dbSNP/1456786974
- dbSNP/1456786975
- dbSNP/1456786976
- dbSNP/1456786977
- dbSNP/1456786978
- dbSNP/1456786979
- dbSNP/1456786980
- dbSNP/1456786981
- dbSNP/1456786982
- dbSNP/1456786983
- dbSNP/1456786984
- dbSNP/1456786985
- dbSNP/1456786986
- dbSNP/1456786987
- dbSNP/1456786988
- dbSNP/1456786989
- dbSNP/1456786990
- dbSNP/1456786991
- dbSNP/1456786992
- dbSNP/1456786993
- dbSNP/1456786994
- dbSNP/1456786995
- dbSNP/1456786996
- dbSNP/1456786997
- dbSNP/1456786998
- dbSNP/1456786999
- dbSNP/1456787000
- dbSNP/1456787001
- dbSNP/1456787002
- dbSNP/1456787003
- dbSNP/1456787004
- dbSNP/1456787005
- dbSNP/1456787006
- dbSNP/1456787007
- dbSNP/1456787008
- dbSNP/1456787009
- dbSNP/1456787010
- dbSNP/1456787011
- dbSNP/1456787012
- dbSNP/1456787013
- dbSNP/1456787014
- dbSNP/1456787015
- dbSNP/1456787016
- dbSNP/1456787017
- dbSNP/1456787018
- dbSNP/1456787019
- dbSNP/1456787020
- dbSNP/1456787021
- dbSNP/1456787022
- dbSNP/1456787023
- dbSNP/1456787024
- dbSNP/1456787025
- dbSNP/1456787026
- dbSNP/1456787027
- dbSNP/1456787028
- dbSNP/1456787029
- dbSNP/1456787030
- dbSNP/1456787031
- dbSNP/1456787032
- dbSNP/1456787033
- dbSNP/1456787034
- dbSNP/1456787035
- dbSNP/1456787036
- dbSNP/1456787037
- dbSNP/1456787038
- dbSNP/1456787039
- dbSNP/1456787040
- dbSNP/1456787041
- dbSNP/1456787042
- dbSNP/1456787043
- dbSNP/1456787044
- dbSNP/1456787045
- dbSNP/1456787046
- dbSNP/1456787047
- dbSNP/1456787048
- dbSNP/1456787049
- dbSNP/1456787050
- dbSNP/1456787051
- dbSNP/1456787052
- dbSNP/1456787053
- dbSNP/1456787054
- dbSNP/1456787055
- dbSNP/1456787056
- dbSNP/1456787057
- dbSNP/1456787058
- dbSNP/1456787059
- dbSNP/1456787060
- dbSNP/1456787061
- dbSNP/1456787062
- dbSNP/1456787063
- dbSNP/1456787064
- dbSNP/1456787065
- dbSNP/1456787066
- dbSNP/1456787067
- dbSNP/1456787068
- dbSNP/1456787069
- dbSNP/1456787070
- dbSNP/1456787071
- dbSNP/1456787072
- dbSNP/1456787073
- dbSNP/1456787074
- dbSNP/1456787075
- dbSNP/1456787076
- dbSNP/1456787077
- dbSNP/1456787078
- dbSNP/1456787079
- dbSNP/1456787080
- dbSNP/1456787081
- dbSNP/1456787082
- dbSNP/1456787083
- dbSNP/1456787084
- dbSNP/1456787085
- dbSNP/1456787086
- dbSNP/1456787087
- dbSNP/1456787088
- dbSNP/1456787089
- dbSNP/1456787090
- dbSNP/1456787091
- dbSNP/1456787092
- dbSNP/1456787093
- dbSNP/1456787094
- dbSNP/1456787095
- dbSNP/1456787096
- dbSNP/1456787097
- dbSNP/1456787098
- dbSNP/1456787099
- dbSNP/1456787100
- dbSNP/1456787101
- dbSNP/1456787102
- dbSNP/1456787103
- dbSNP/1456787104
- dbSNP/1456787105
- dbSNP/1456787106
- dbSNP/1456787107
- dbSNP/1456787108
- dbSNP/1456787109
- dbSNP/1456787110
- dbSNP/1456787111
- dbSNP/1456787112
- dbSNP/1456787113
- dbSNP/1456787114
- dbSNP/1456787115
- dbSNP/1456787116
- dbSNP/1456787117
- dbSNP/1456787118
- dbSNP/1456787119
- dbSNP/1456787120
- dbSNP/1456787121
- dbSNP/1456787122
- dbSNP/1456787123
- dbSNP/1456787124
- dbSNP/1456787125
- dbSNP/1456787126
- dbSNP/1456787127
- dbSNP/1456787128
- dbSNP/1456787129
- dbSNP/1456787130
- dbSNP/1456787131
- dbSNP/1456787132
- dbSNP/1456787133
- dbSNP/1456787134
- dbSNP/1456787135
- dbSNP/1456787136
- dbSNP/1456787137
- dbSNP/1456787138
- dbSNP/1456787139
- dbSNP/1456787140
- dbSNP/1456787141
- dbSNP/1456787142
- dbSNP/1456787143
- dbSNP/1456787144
- dbSNP/1456787145
- dbSNP/1456787146
- dbSNP/1456787147
- dbSNP/1456787148
- dbSNP/1456787149
- dbSNP/1456787150
- dbSNP/1456787151
- dbSNP/1456787152
- dbSNP/1456787153
- dbSNP/1456787154
- dbSNP/1456787155
- dbSNP/1456787156
- dbSNP/1456787157
- dbSNP/1456787158
- dbSNP/1456787159
- dbSNP/1456787160
- dbSNP/1456787161
- dbSNP/1456787162
- dbSNP/1456787163
- dbSNP/1456787164
- dbSNP/1456787165
- dbSNP/1456787166
- dbSNP/1456787167
- dbSNP/1456787168
- dbSNP/1456787169
- dbSNP/1456787170
- dbSNP/1456787171
- dbSNP/1456787172
- dbSNP/1456787173
- dbSNP/1456787174
- dbSNP/1456787175
- dbSNP/1456787176
- dbSNP/1456787177
- dbSNP/1456787178
- dbSNP/1456787179
- dbSNP/1456787180
- dbSNP/1456787181
- dbSNP/1456787182
- dbSNP/1456787183
- dbSNP/1456787184
- dbSNP/1456787185
- dbSNP/1456787186
- dbSNP/1456787187
- dbSNP/1456787188
- dbSNP/1456787189
- dbSNP/1456787190
- dbSNP/1456787191
- dbSNP/1456787192
- dbSNP/1456787193
- dbSNP/1456787194
- dbSNP/1456787195
- dbSNP/1456787196
- dbSNP/1456787197
- dbSNP/1456787198
- dbSNP/1456787199
- dbSNP/1456787200
- dbSNP/1456787201
- dbSNP/1456787202
- dbSNP/1456787203
- dbSNP/1456787204
- dbSNP/1456787205
- dbSNP/1456787206
- dbSNP/1456787207
- dbSNP/1456787208
- dbSNP/1456787209
- dbSNP/1456787210
- dbSNP/1456787211
- dbSNP/1456787212
- dbSNP/1456787213
- dbSNP/1456787214
- dbSNP/1456787215
- dbSNP/1456787216
- dbSNP/1456787217
- dbSNP/1456787218
- dbSNP/1456787219
- dbSNP/1456787220
- dbSNP/1456787221
- dbSNP/1456787222
- dbSNP/1456787223
- dbSNP/1456787224
- dbSNP/1456787225
- dbSNP/1456787226
- dbSNP/1456787227
- dbSNP/1456787228
- dbSNP/1456787229
- dbSNP/1456787230
- dbSNP/1456787231
- dbSNP/1456787232
- dbSNP/1456787233
- dbSNP/1456787234
- dbSNP/1456787235
- dbSNP/1456787236
- dbSNP/1456787237
- dbSNP/1456787238
- dbSNP/1456787239
- dbSNP/1456787240
- dbSNP/1456787241
- dbSNP/1456787242
- dbSNP/1456787243
- dbSNP/1456787244
- dbSNP/1456787245
- dbSNP/1456787246
- dbSNP/1456787247
- dbSNP/1456787248
- dbSNP/1456787249
- dbSNP/1456787250
- dbSNP/1456787251
- dbSNP/1456787252
- dbSNP/1456787253
- dbSNP/1456787254
- dbSNP/1456787255
- dbSNP/1456787256
- dbSNP/1456787257
- dbSNP/1456787258
- dbSNP/1456787259
- dbSNP/1456787260
- dbSNP/1456787261
- dbSNP/1456787262
- dbSNP/1456787263
- dbSNP/1456787264
- dbSNP/1456787265
- dbSNP/1456787266
- dbSNP/1456787267
- dbSNP/1456787268
- dbSNP/1456787269
- dbSNP/1456787270
- dbSNP/1456787271
- dbSNP/1456787272
- dbSNP/1456787273
- dbSNP/1456787274
- dbSNP/1456787275
Grants and funding
- HHSN272200900018C/AI/NIAID NIH HHS/United States
- AI081704/AI/NIAID NIH HHS/United States
- R21 AI112363/AI/NIAID NIH HHS/United States
- AI0624273/AI/NIAID NIH HHS/United States
- R01 AI075096/AI/NIAID NIH HHS/United States
- HHSN272200900018C/PHS HHS/United States
- AI075096-S1/AI/NIAID NIH HHS/United States
- F31DE023726/DE/NIDCR NIH HHS/United States
- F31 DE023726/DE/NIDCR NIH HHS/United States
- AI075096/AI/NIAID NIH HHS/United States
- AI112363/AI/NIAID NIH HHS/United States
- U19 AI110818/AI/NIAID NIH HHS/United States
- R01 AI081704/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
