Engineering high-potency R-spondin adult stem cell growth factors
- PMID: 25504990
- PMCID: PMC4352588
- DOI: 10.1124/mol.114.095133
Engineering high-potency R-spondin adult stem cell growth factors
Abstract
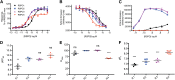
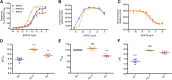
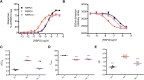
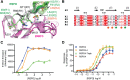
Secreted R-spondin proteins (RSPOs1-4) function as adult stem cell growth factors by potentiating Wnt signaling. Simultaneous binding of distinct regions of the RSPO Fu1-Fu2 domain module to the extracellular domains (ECDs) of the LGR4 G protein-coupled receptor and the ZNRF3 transmembrane E3 ubiquitin ligase regulates Wnt receptor availability. Here, we examine the molecular basis for the differing signaling strengths of RSPOs1-4 using purified RSPO Fu1-Fu2, LGR4 ECD, and ZNRF3 ECD proteins in Wnt signaling and receptor binding assays, and we engineer novel high-potency RSPOs. RSPO2/3/4 had similar signaling potencies that were stronger than that of RSPO1, whereas RSPO1/2/3 had similar efficacies that were greater than that of RSPO4. The RSPOs bound LGR4 with affinity rank order RSPO4 > RSPO2/3 > RSPO1 and ZNRF3 with affinity rank order RSPO2/3 > > RSPO1 > RSPO4. An RSPO2-4 chimera combining RSPO2 ZNRF3 binding with RSPO4 LGR4 binding was a "Superspondin" that exhibited enhanced ternary complex formation and 10-fold stronger signaling potency than RSPO2 and efficacy equivalent to RSPO2. An RSPO4-1 chimera combining RSPO4 ZNRF3 binding with RSPO1 LGR4 binding was a "Poorspondin" that exhibited signaling potency similar to RSPO1 and efficacy equivalent to RSPO4. Conferring increased ZNRF3 binding upon RSPO4 with amino acid substitutions L56F, I58L, and I63M enhanced its signaling potency and efficacy. Our results reveal the molecular basis for RSPOs1-4 activity differences and suggest that signaling potency is determined by ternary complex formation ability, whereas efficacy depends on ZNRF3 recruitment. High-potency RSPOs may be of value for regenerative medicine and/or therapeutic applications.
Copyright © 2015 by The American Society for Pharmacology and Experimental Therapeutics.
Figures




References
-
- Aoki M, Mieda M, Ikeda T, Hamada Y, Nakamura H, Okamoto H. (2007) R-spondin3 is required for mouse placental development. Dev Biol 301:218–226. - PubMed
-
- Aricescu AR, Lu W, Jones EY. (2006) A time- and cost-efficient system for high-level protein production in mammalian cells. Acta Crystallogr D Biol Crystallogr 62:1243–1250. - PubMed
-
- Barker N, Clevers H. (2010) Leucine-rich repeat-containing G-protein-coupled receptors as markers of adult stem cells. Gastroenterology 138:1681–1696. - PubMed
-
- Bell SM, Schreiner CM, Wert SE, Mucenski ML, Scott WJ, Whitsett JA. (2008) R-spondin 2 is required for normal laryngeal-tracheal, lung and limb morphogenesis. Development 135:1049–1058. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

