MicroRNA-mediated transformation by the Kaposi's sarcoma-associated herpesvirus Kaposin locus
- PMID: 25505059
- PMCID: PMC4338870
- DOI: 10.1128/JVI.03317-14
MicroRNA-mediated transformation by the Kaposi's sarcoma-associated herpesvirus Kaposin locus
Abstract
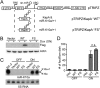
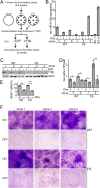
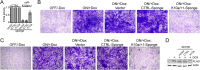
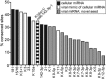
The human oncogenic Kaposi's sarcoma-associated herpesvirus (KSHV) expresses a set of ∼20 viral microRNAs (miRNAs). miR-K10a stands out among these miRNAs because its entire stem-loop precursor overlaps the coding sequence for the Kaposin (Kap) A/C proteins. The ectopic expression of KapA has been reported to lead to transformation of rodent fibroblasts. However, these experiments inadvertently also introduced miR-K10a, which raises the question whether the transforming activity of the locus could in fact be due to miR-K10a expression. To answer this question, we have uncoupled miR-K10a and KapA expression. Our experiments revealed that miR-K10a alone transformed cells with an efficiency similar to that when it was coexpressed with KapA. Maintenance of the transformed phenotype was conditional upon continued miR-K10a but not KapA protein expression, consistent with its dependence on miRNA-mediated changes in gene expression. Importantly, miR-K10a taps into an evolutionarily conserved network of miR-142-3p targets, several of which are expressed in 3T3 cells and are also known inhibitors of cellular transformation. In summary, our studies of miR-K10a serve as an example of an unsuspected function of an mRNA whose precursor is embedded within a coding transcript. In addition, our identification of conserved miR-K10a targets that limit transformation will point the way to a better understanding of the role of this miRNA in KSHV-associated tumors.
Importance: Kaposi's sarcoma-associated herpesvirus (KSHV) is a human tumor virus. The viral Kaposin locus has known oncogenic potential, which has previously been attributed to the encoded KapA protein. Here we show that the virally encoded miR-K10a miRNA, whose precursor overlaps the KapA-coding region, may account for the oncogenic properties of this locus. Our data suggest that miR-K10a mimics the cellular miRNA miR-142-3p and thereby represses several known inhibitors of oncogenic transformation. Our work demonstrates that functional properties attributed to a coding region may in fact be carried out by an embedded noncoding element and sheds light on the functions of viral miR-K10a.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures


Similar articles
-
Regulation of tumor necrosis factor-like weak inducer of apoptosis receptor protein (TWEAKR) expression by Kaposi's sarcoma-associated herpesvirus microRNA prevents TWEAK-induced apoptosis and inflammatory cytokine expression.J Virol. 2010 Dec;84(23):12139-51. doi: 10.1128/JVI.00884-10. Epub 2010 Sep 15. J Virol. 2010. PMID: 20844036 Free PMC article.
-
A Panel of Kaposi's Sarcoma-Associated Herpesvirus Mutants in the Polycistronic Kaposin Locus for Precise Analysis of Individual Protein Products.J Virol. 2022 Mar 9;96(5):e0156021. doi: 10.1128/JVI.01560-21. Epub 2021 Dec 22. J Virol. 2022. PMID: 34936820 Free PMC article.
-
Modified Cross-Linking, Ligation, and Sequencing of Hybrids (qCLASH) Identifies Kaposi's Sarcoma-Associated Herpesvirus MicroRNA Targets in Endothelial Cells.J Virol. 2018 Mar 28;92(8):e02138-17. doi: 10.1128/JVI.02138-17. Print 2018 Apr 15. J Virol. 2018. PMID: 29386283 Free PMC article.
-
Posttranscriptional gene regulation in Kaposi's sarcoma-associated herpesvirus.Adv Appl Microbiol. 2009;68:241-61. doi: 10.1016/S0065-2164(09)01206-4. Adv Appl Microbiol. 2009. PMID: 19426857 Review.
-
Regulation of the MIR155 host gene in physiological and pathological processes.Gene. 2013 Dec 10;532(1):1-12. doi: 10.1016/j.gene.2012.12.009. Epub 2012 Dec 14. Gene. 2013. PMID: 23246696 Review.
Cited by
-
Identification of the X-linked germ cell specific miRNAs (XmiRs) and their functions.PLoS One. 2019 Feb 1;14(2):e0211739. doi: 10.1371/journal.pone.0211739. eCollection 2019. PLoS One. 2019. PMID: 30707741 Free PMC article.
-
Genomic changes in Kaposi Sarcoma-associated Herpesvirus and their clinical correlates.PLoS Pathog. 2022 Nov 28;18(11):e1010524. doi: 10.1371/journal.ppat.1010524. eCollection 2022 Nov. PLoS Pathog. 2022. PMID: 36441790 Free PMC article.
-
Kaposi's sarcoma herpesvirus (KSHV) microRNA K12-1 functions as an oncogene by activating NF-κB/IL-6/STAT3 signaling.Oncotarget. 2016 May 31;7(22):33363-73. doi: 10.18632/oncotarget.9221. Oncotarget. 2016. PMID: 27166260 Free PMC article.
-
The cellular and KSHV A-to-I RNA editome in primary effusion lymphoma and its role in the viral lifecycle.Nat Commun. 2023 Mar 13;14(1):1367. doi: 10.1038/s41467-023-37105-8. Nat Commun. 2023. PMID: 36914661 Free PMC article.
-
Herpesvirus microRNAs for use in gene therapy immune-evasion strategies.Gene Ther. 2017 Jul;24(7):385-391. doi: 10.1038/gt.2017.37. Epub 2017 May 9. Gene Ther. 2017. PMID: 28485720 Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

