Identification and functional comparison of seven-transmembrane G-protein-coupled BILF1 receptors in recently discovered nonhuman primate lymphocryptoviruses
- PMID: 25505061
- PMCID: PMC4338891
- DOI: 10.1128/JVI.02716-14
Identification and functional comparison of seven-transmembrane G-protein-coupled BILF1 receptors in recently discovered nonhuman primate lymphocryptoviruses
Abstract
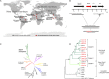
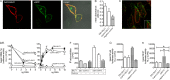
Coevolution of herpesviruses with their respective host has resulted in a delicate balance between virus-encoded immune evasion mechanisms and host antiviral immunity. BILF1 encoded by human Epstein-Barr virus (EBV) is a 7-transmembrane (7TM) G-protein-coupled receptor (GPCR) with multiple immunomodulatory functions, including attenuation of PKR phosphorylation, activation of G-protein signaling, and downregulation of major histocompatibility complex (MHC) class I surface expression. In this study, we explored the evolutionary and functional relationships between BILF1 receptor family members from EBV and 12 previously uncharacterized nonhuman primate (NHP) lymphocryptoviruses (LCVs). Phylogenetic analysis defined 3 BILF1 clades, corresponding to LCVs of New World monkeys (clade A) or Old World monkeys and great apes (clades B and C). Common functional properties were suggested by a high degree of sequence conservation in functionally important regions of the BILF1 molecules. A subset of BILF1 receptors from EBV and LCVs from NHPs (chimpanzee, orangutan, marmoset, and siamang) were selected for multifunctional analysis. All receptors exhibited constitutive signaling activity via G protein Gαi and induced activation of the NF-κB transcription factor. In contrast, only 3 of 5 were able to activate NFAT (nuclear factor of activated T cells); chimpanzee and orangutan BILF1 molecules were unable to activate NFAT. Similarly, although all receptors were internalized, BILF1 from the chimpanzee and orangutan displayed an altered cellular localization pattern with predominant cell surface expression. This study shows how biochemical characterization of functionally important orthologous viral proteins can be used to complement phylogenetic analysis to provide further insight into diverse microbial evolutionary relationships and immune evasion function.
Importance: Epstein-Barr virus (EBV), known as an oncovirus, is the only human herpesvirus in the genus Lymphocryptovirus (LCV). EBV uses multiple strategies to hijack infected host cells, establish persistent infection in B cells, and evade antiviral immune responses. As part of EBV's immune evasion strategy, the virus encodes a multifunctional 7-transmembrane (7TM) G-protein-coupled receptor (GPCR), EBV BILF1. In addition to multiple immune evasion-associated functions, EBV BILF1 has transforming properties, which are linked to its high constitutive activity. We identified BILF1 receptor orthologues in 12 previously uncharacterized LCVs from nonhuman primates (NHPs) of Old and New World origin. As 7TM receptors are excellent drug targets, our unique insight into the molecular mechanism of action of the BILF1 family and into the evolution of primate LCVs may enable validation of EBV BILF1 as a drug target for EBV-mediated diseases, as well as facilitating the design of drugs targeting EBV BILF1.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures





Similar articles
-
Distinct Roles of Extracellular Domains in the Epstein-Barr Virus-Encoded BILF1 Receptor for Signaling and Major Histocompatibility Complex Class I Downregulation.mBio. 2019 Jan 15;10(1):e01707-18. doi: 10.1128/mBio.01707-18. mBio. 2019. PMID: 30647152 Free PMC article.
-
The Epstein-Barr virus G-protein-coupled receptor contributes to immune evasion by targeting MHC class I molecules for degradation.PLoS Pathog. 2009 Jan;5(1):e1000255. doi: 10.1371/journal.ppat.1000255. Epub 2009 Jan 2. PLoS Pathog. 2009. PMID: 19119421 Free PMC article.
-
Complete genomic sequence of an Epstein-Barr virus-related herpesvirus naturally infecting a new world primate: a defining point in the evolution of oncogenic lymphocryptoviruses.J Virol. 2002 Dec;76(23):12055-68. doi: 10.1128/jvi.76.23.12055-12068.2002. J Virol. 2002. PMID: 12414947 Free PMC article.
-
Non-human Primate Lymphocryptoviruses: Past, Present, and Future.Curr Top Microbiol Immunol. 2015;391:385-405. doi: 10.1007/978-3-319-22834-1_13. Curr Top Microbiol Immunol. 2015. PMID: 26428382 Review.
-
EBV, the human host, and the 7TM receptors: defense or offense?Prog Mol Biol Transl Sci. 2015;129:395-427. doi: 10.1016/bs.pmbts.2014.10.011. Epub 2014 Dec 1. Prog Mol Biol Transl Sci. 2015. PMID: 25595811 Review.
Cited by
-
Manipulation of the host cell membrane by human γ-herpesviruses EBV and KSHV for pathogenesis.Virol Sin. 2016 Oct;31(5):395-405. doi: 10.1007/s12250-016-3817-2. Epub 2016 Sep 12. Virol Sin. 2016. PMID: 27624182 Free PMC article. Review.
-
Patterns of human and porcine gammaherpesvirus-encoded BILF1 receptor endocytosis.Cell Mol Biol Lett. 2023 Feb 21;28(1):14. doi: 10.1186/s11658-023-00427-y. Cell Mol Biol Lett. 2023. PMID: 36810008 Free PMC article.
-
Methods for Studying Endocytotic Pathways of Herpesvirus Encoded G Protein-Coupled Receptors.Molecules. 2020 Dec 3;25(23):5710. doi: 10.3390/molecules25235710. Molecules. 2020. PMID: 33287269 Free PMC article. Review.
-
Molecular Properties and Therapeutic Targeting of the EBV-Encoded Receptor BILF1.Cancers (Basel). 2021 Aug 13;13(16):4079. doi: 10.3390/cancers13164079. Cancers (Basel). 2021. PMID: 34439235 Free PMC article. Review.
-
Specifically Targeted Transport of Plasma Membrane Transporters: From Potential Mechanisms for Regulating Cell Health or Disease to Applications.Membranes (Basel). 2021 Sep 27;11(10):736. doi: 10.3390/membranes11100736. Membranes (Basel). 2021. PMID: 34677502 Free PMC article. Review.
References
-
- Hislop AD, Ressing ME, van Leeuwen D, Pudney VA, Horst D, Koppers-Lalic D, Croft NP, Neefjes JJ, Rickinson AB, Wiertz EJHJ. 2007. A CD8+ T cell immune evasion protein specific to Epstein-Barr virus and its close relatives in Old World primates. J Exp Med 204:1863–1873. doi:10.1084/jem.20070256. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

