The nucleoprotein of newly emerged H7N9 influenza A virus harbors a unique motif conferring resistance to antiviral human MxA
- PMID: 25505067
- PMCID: PMC4338896
- DOI: 10.1128/JVI.02406-14
The nucleoprotein of newly emerged H7N9 influenza A virus harbors a unique motif conferring resistance to antiviral human MxA
Abstract
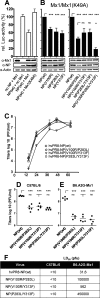
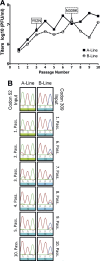
Interferon-induced Mx proteins show strong antiviral activity against influenza A viruses (IAVs). We recently demonstrated that the viral nucleoprotein (NP) determines resistance of seasonal and pandemic human influenza viruses to Mx, while avian isolates retain Mx sensitivity. We identified a surface-exposed cluster of amino acids in NP of pandemic A/BM/1/1918 (H1N1), comprising isoleucine-100, proline-283, and tyrosine-313, that is essential for reduced Mx sensitivity in cell culture and in vivo. This cluster has been maintained in all descendant seasonal strains, including A/PR/8/34 (PR/8). Accordingly, two substitutions in the NP of PR/8 [PR/8(mut)] to the Mx-sensitive amino acids (P283L and Y313F) led to attenuation in Mx1-positive mice. Serial lung passages of PR/8(mut) in Mx1 mice resulted in a single exchange of tyrosine to asparagine at position 52 in NP (in close proximity to the amino acid cluster at positions 100, 283, and 313), which partially compensates loss of Mx resistance in PR/8(mut). Intriguingly, the NP of the newly emerged avian-origin H7N9 virus also contains an asparagine at position 52 and shows reduced Mx sensitivity. N52Y substitution in NP results in increased sensitivity of the H7N9 virus to human Mx, indicating that this residue is a determinant of Mx resistance in mammals. Our data strengthen the hypothesis that the human Mx protein represents a potent barrier against zoonotic transmission of avian influenza viruses. However, the H7N9 viruses overcome this restriction by harboring an NP that is less sensitive to Mx-mediated host defense. This might contribute to zoonotic transmission of H7N9 and to the severe to fatal outcome of H7N9 infections in humans.
Importance: The natural host of influenza A viruses (IAVs) are aquatic birds. Occasionally, these viruses cross the species barrier, as in early 2013 when an avian H7N9 virus infected humans in China. Since then, multiple transmissions of H7N9 viruses to humans have occurred, leaving experts puzzled about molecular causes for such efficient crossing of the species barrier compared to other avian influenza viruses. Mx proteins are known restriction factors preventing influenza virus replication. Unfortunately, some viruses (e.g., human IAV) have developed some resistance, which is associated with specific amino acids in their nucleoproteins, the target of Mx function. Here, we demonstrate that the novel H7N9 bird IAV already carries a nucleoprotein that overcomes the inhibition of viral replication by human MxA. This is the first example of an avian IAV that is naturally less sensitive to Mx-mediated inhibition and might explain why H7N9 viruses transmitted efficiently to humans.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures
Similar articles
-
Eurasian Avian-Like Swine Influenza A Viruses Escape Human MxA Restriction through Distinct Mutations in Their Nucleoprotein.J Virol. 2019 Jan 4;93(2):e00997-18. doi: 10.1128/JVI.00997-18. J Virol. 2019. PMID: 30355693 Free PMC article.
-
The viral nucleoprotein determines Mx sensitivity of influenza A viruses.J Virol. 2011 Aug;85(16):8133-40. doi: 10.1128/JVI.00712-11. Epub 2011 Jun 15. J Virol. 2011. PMID: 21680506 Free PMC article.
-
H5N1 Influenza A Virus PB1-F2 Relieves HAX-1-Mediated Restriction of Avian Virus Polymerase PA in Human Lung Cells.J Virol. 2018 May 14;92(11):e00425-18. doi: 10.1128/JVI.00425-18. Print 2018 Jun 1. J Virol. 2018. PMID: 29563290 Free PMC article.
-
Pandemic potential of avian influenza A (H7N9) viruses.Trends Microbiol. 2014 Nov;22(11):623-31. doi: 10.1016/j.tim.2014.08.008. Epub 2014 Sep 25. Trends Microbiol. 2014. PMID: 25264312 Free PMC article. Review.
-
The Pandemic Threat of Emerging H5 and H7 Avian Influenza Viruses.Viruses. 2018 Aug 28;10(9):461. doi: 10.3390/v10090461. Viruses. 2018. PMID: 30154345 Free PMC article. Review.
Cited by
-
The Nucleoprotein of H7N9 Influenza Virus Positively Regulates TRAF3-Mediated Innate Signaling and Attenuates Viral Virulence in Mice.J Virol. 2020 Nov 23;94(24):e01640-20. doi: 10.1128/JVI.01640-20. Print 2020 Nov 23. J Virol. 2020. PMID: 33028715 Free PMC article.
-
Zoonotic Potential of Influenza A Viruses: A Comprehensive Overview.Viruses. 2018 Sep 13;10(9):497. doi: 10.3390/v10090497. Viruses. 2018. PMID: 30217093 Free PMC article. Review.
-
One health, multiple challenges: The inter-species transmission of influenza A virus.One Health. 2015 Dec 1;1:1-13. doi: 10.1016/j.onehlt.2015.03.001. One Health. 2015. PMID: 26309905 Free PMC article.
-
Sphingosine 1-Phosphate Lyase Enhances the Activation of IKKε To Promote Type I IFN-Mediated Innate Immune Responses to Influenza A Virus Infection.J Immunol. 2017 Jul 15;199(2):677-687. doi: 10.4049/jimmunol.1601959. Epub 2017 Jun 9. J Immunol. 2017. PMID: 28600291 Free PMC article.
-
The ecology and adaptive evolution of influenza A interspecies transmission.Influenza Other Respir Viruses. 2017 Jan;11(1):74-84. doi: 10.1111/irv.12412. Epub 2016 Aug 8. Influenza Other Respir Viruses. 2017. PMID: 27426214 Free PMC article. Review.
References
-
- Chen Y, Liang W, Yang S, Wu N, Gao H, Sheng J, Yao H, Wo J, Fang Q, Cui D, Li Y, Yao X, Zhang Y, Wu H, Zheng S, Diao H, Xia S, Zhang Y, Chan KH, Tsoi HW, Teng JL, Song W, Wang P, Lau SY, Zheng M, Chan JF, To KK, Chen H, Li L, Yuen KY. 2013. Human infections with the emerging avian influenza A H7N9 virus from wet market poultry: clinical analysis and characterisation of viral genome. Lancet 381:1916–1925. doi:10.1016/S0140-6736(13)60903-4. - DOI - PMC - PubMed
-
- Gao R, Cao B, Hu Y, Feng Z, Wang D, Hu W, Chen J, Jie Z, Qiu H, Xu K, Xu X, Lu H, Zhu W, Gao Z, Xiang N, Shen Y, He Z, Gu Y, Zhang Z, Yang Y, Zhao X, Zhou L, Li X, Zou S, Zhang Y, Li X, Yang L, Guo J, Dong J, Li Q, Dong L, Zhu Y, Bai T, Wang S, Hao P, Yang W, Zhang Y, Han J, Yu H, Li D, Gao GF, Wu G, Wang Y, Yuan Z, Shu Y. 2013. Human infection with a novel avian-origin influenza A (H7N9) virus. N Engl J Med 368:1888–1897. doi:10.1056/NEJMoa1304459. - DOI - PubMed
-
- Kageyama T, Fujisaki S, Takashita E, Xu H, Yamada S, Uchida Y, Neumann G, Saito T, Kawaoka Y, Tashiro M. 2013. Genetic analysis of novel avian A(H7N9) influenza viruses isolated from patients in China, February to April 2013. Euro Surveill 18:20453 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=20453. - PMC - PubMed
-
- Zhang Q, Shi J, Deng G, Guo J, Zeng X, He X, Kong H, Gu C, Li X, Liu J, Wang G, Chen Y, Liu L, Liang L, Li Y, Fan J, Wang J, Li W, Guan L, Li Q, Yang H, Chen P, Jiang L, Guan Y, Xin X, Jiang Y, Tian G, Wang X, Qiao C, Li C, Bu Z, Chen H. 2013. H7N9 influenza viruses are transmissible in ferrets by respiratory droplet. Science 341:410–414. doi:10.1126/science.1240532. - DOI - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

