Topological analysis of Hedgehog acyltransferase, a multipalmitoylated transmembrane protein
- PMID: 25505265
- PMCID: PMC4319003
- DOI: 10.1074/jbc.M114.614578
Topological analysis of Hedgehog acyltransferase, a multipalmitoylated transmembrane protein
Abstract
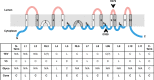
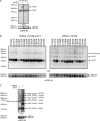
Hedgehog proteins are secreted morphogens that play critical roles in development and disease. During maturation of the proteins through the secretory pathway, they are modified by the addition of N-terminal palmitic acid and C-terminal cholesterol moieties, both of which are critical for their correct function and localization. Hedgehog acyltransferase (HHAT) is the enzyme in the endoplasmic reticulum that palmitoylates Hedgehog proteins, is a member of a small subfamily of membrane-bound O-acyltransferase proteins that acylate secreted proteins, and is an important drug target in cancer. However, little is known about HHAT structure and mode of function. We show that HHAT is comprised of ten transmembrane domains and two reentrant loops with the critical His and Asp residues on opposite sides of the endoplasmic reticulum membrane. We further show that HHAT is palmitoylated on multiple cytosolic cysteines that maintain protein structure within the membrane. Finally, we provide evidence that mutation of the conserved His residue in the hypothesized catalytic domain results in a complete loss of HHAT palmitoylation, providing novel insights into how the protein may function in vivo.
Keywords: Acyltransferase; Cancer; Click Chemistry; HHAT; Hedgehog Signaling Pathway; MBOAT; Protein Palmitoylation; Topology; Transmembrane Domain.
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures


Similar articles
-
Membrane topology of hedgehog acyltransferase.J Biol Chem. 2015 Jan 23;290(4):2235-43. doi: 10.1074/jbc.M114.625764. Epub 2014 Dec 8. J Biol Chem. 2015. PMID: 25488661 Free PMC article.
-
Palmitoylation of Hedgehog proteins by Hedgehog acyltransferase: roles in signalling and disease.Open Biol. 2021 Mar;11(3):200414. doi: 10.1098/rsob.200414. Epub 2021 Mar 3. Open Biol. 2021. PMID: 33653085 Free PMC article. Review.
-
Click chemistry armed enzyme-linked immunosorbent assay to measure palmitoylation by hedgehog acyltransferase.Anal Biochem. 2015 Dec 1;490:66-72. doi: 10.1016/j.ab.2015.08.025. Epub 2015 Sep 1. Anal Biochem. 2015. PMID: 26334609 Free PMC article.
-
Substrate and product complexes reveal mechanisms of Hedgehog acylation by HHAT.Science. 2021 Jun 11;372(6547):1215-1219. doi: 10.1126/science.abg4998. Science. 2021. PMID: 34112694 Free PMC article.
-
Palmitoylation of Hedgehog proteins.Vitam Horm. 2012;88:229-52. doi: 10.1016/B978-0-12-394622-5.00010-9. Vitam Horm. 2012. PMID: 22391306 Free PMC article. Review.
Cited by
-
Chemogenetic E-MAP in Saccharomyces cerevisiae for Identification of Membrane Transporters Operating Lipid Flip Flop.PLoS Genet. 2016 Jul 27;12(7):e1006160. doi: 10.1371/journal.pgen.1006160. eCollection 2016 Jul. PLoS Genet. 2016. PMID: 27462707 Free PMC article.
-
Determination of the membrane topology of PORCN, an O-acyl transferase that modifies Wnt signalling proteins.Open Biol. 2021 Jun;11(6):200400. doi: 10.1098/rsob.200400. Epub 2021 Jun 30. Open Biol. 2021. PMID: 34186010 Free PMC article.
-
Hedgehog Acyltransferase Promotes Uptake of Palmitoyl-CoA across the Endoplasmic Reticulum Membrane.Cell Rep. 2019 Dec 24;29(13):4608-4619.e4. doi: 10.1016/j.celrep.2019.11.110. Cell Rep. 2019. PMID: 31875564 Free PMC article.
-
Yeast Gup1(2) Proteins Are Homologues of the Hedgehog Morphogens Acyltransferases HHAT(L): Facts and Implications.J Dev Biol. 2016 Nov 5;4(4):33. doi: 10.3390/jdb4040033. J Dev Biol. 2016. PMID: 29615596 Free PMC article. Review.
-
A rising tide lifts all MBOATs: recent progress in structural and functional understanding of membrane bound O-acyltransferases.Front Physiol. 2023 May 4;14:1167873. doi: 10.3389/fphys.2023.1167873. eCollection 2023. Front Physiol. 2023. PMID: 37250116 Free PMC article. Review.
References
-
- McMahon A. P., Ingham P. W., Tabin C. J. (2003) Developmental roles and clinical significance of hedgehog signaling. Curr. Top. Dev. Biol. 53, 1–114 - PubMed
-
- Ingham P. W., McMahon A. P. (2001) Hedgehog signaling in animal development: paradigms and principles. Genes Dev. 15, 3059–3087 - PubMed
-
- Scales S. J., de Sauvage F. J. (2009) Mechanisms of Hedgehog pathway activation in cancer and implications for therapy. Trends Pharmacol. Sci. 30, 303–312 - PubMed
-
- Mann R. K., Beachy P. A. (2004) Novel lipid modifications of secreted protein signals. Annu. Rev. Biochem. 73, 891–923 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

