Identification and nomenclature of the genus Penicillium
- PMID: 25505353
- PMCID: PMC4261876
- DOI: 10.1016/j.simyco.2014.09.001
Identification and nomenclature of the genus Penicillium
Abstract
Penicillium is a diverse genus occurring worldwide and its species play important roles as decomposers of organic materials and cause destructive rots in the food industry where they produce a wide range of mycotoxins. Other species are considered enzyme factories or are common indoor air allergens. Although DNA sequences are essential for robust identification of Penicillium species, there is currently no comprehensive, verified reference database for the genus. To coincide with the move to one fungus one name in the International Code of Nomenclature for algae, fungi and plants, the generic concept of Penicillium was re-defined to accommodate species from other genera, such as Chromocleista, Eladia, Eupenicillium, Torulomyces and Thysanophora, which together comprise a large monophyletic clade. As a result of this, and the many new species described in recent years, it was necessary to update the list of accepted species in Penicillium. The genus currently contains 354 accepted species, including new combinations for Aspergillus crystallinus, A. malodoratus and A. paradoxus, which belong to Penicillium section Paradoxa. To add to the taxonomic value of the list, we also provide information on each accepted species MycoBank number, living ex-type strains and provide GenBank accession numbers to ITS, β-tubulin, calmodulin and RPB2 sequences, thereby supplying a verified set of sequences for each species of the genus. In addition to the nomenclatural list, we recommend a standard working method for species descriptions and identifications to be adopted by laboratories working on this genus.
Keywords: Aspergillaceae; Fungal identification; media; nomenclature; phylogeny.
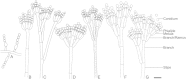
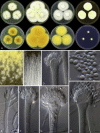
Figures




References
-
- Abraham E., Chain E., Fletcher C. Further observations on penicillin. The Lancet. 1941;238:177–188.
-
- Adsul M.G., Bastawde K.B., Varma A.J. Strain improvement of Penicillium janthinellum NCIM 1171 for increased cellulase production. Bioresource Technology. 2007;98:1467–1473. - PubMed
-
- Amiri-Eliasi B., Fenselau C. Characterization of protein biomarkers desorbed by MALDI from whole fungal cells. Analytical Chemistry. 2001;73:5228–5231. - PubMed
-
- Barreto M.C., Houbraken J., Samson R.A. Taxonomic studies of the Penicillium glabrum complex and the description of a new species P. subericola. Fungal Diversity. 2011;49:23–33.
-
- Basaran P., Demirbas R.M. Spectroscopic detection of pharmaceutical compounds from an aflatoxigenic strain of Aspergillus parasiticus. Microbiological Research. 2010;165:516–522. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
