Mechanisms of polymyxin resistance: acquired and intrinsic resistance in bacteria
- PMID: 25505462
- PMCID: PMC4244539
- DOI: 10.3389/fmicb.2014.00643
Mechanisms of polymyxin resistance: acquired and intrinsic resistance in bacteria
Abstract
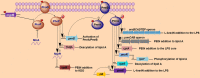
Polymyxins are polycationic antimicrobial peptides that are currently the last-resort antibiotics for the treatment of multidrug-resistant, Gram-negative bacterial infections. The reintroduction of polymyxins for antimicrobial therapy has been followed by an increase in reports of resistance among Gram-negative bacteria. Some bacteria, such as Klebsiella pneumoniae, Pseudomonas aeruginosa, and Acinetobacter baumannii, develop resistance to polymyxins in a process referred to as acquired resistance, whereas other bacteria, such as Proteus spp., Serratia spp., and Burkholderia spp., are naturally resistant to these drugs. Reports of polymyxin resistance in clinical isolates have recently increased, including acquired and intrinsically resistant pathogens. This increase is considered a serious issue, prompting concern due to the low number of currently available effective antibiotics. This review summarizes current knowledge concerning the different strategies bacteria employ to resist the activities of polymyxins. Gram-negative bacteria employ several strategies to protect themselves from polymyxin antibiotics (polymyxin B and colistin), including a variety of lipopolysaccharide (LPS) modifications, such as modifications of lipid A with phosphoethanolamine and 4-amino-4-deoxy-L-arabinose, in addition to the use of efflux pumps, the formation of capsules and overexpression of the outer membrane protein OprH, which are all effectively regulated at the molecular level. The increased understanding of these mechanisms is extremely vital and timely to facilitate studies of antimicrobial peptides and find new potential drugs targeting clinically relevant Gram-negative bacteria.
Keywords: Enterobacteriaceae; antibiotic resistance; lipid A; lipopolysaccharides; mutation; non-fermentative bacilli; two-component systems.
Figures
References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

