Molecular determinants of staphylococcal biofilm dispersal and structuring
- PMID: 25505739
- PMCID: PMC4244807
- DOI: 10.3389/fcimb.2014.00167
Molecular determinants of staphylococcal biofilm dispersal and structuring
Abstract
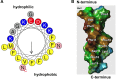
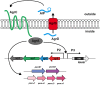
Staphylococci are frequently implicated in human infections, and continue to pose a therapeutic dilemma due to their ability to form deeply seated microbial communities, known as biofilms, on the surfaces of implanted medical devices and host tissues. Biofilm development has been proposed to occur in three stages: (1) attachment, (2) proliferation/structuring, and (3) detachment/dispersal. Although research within the last several decades has implicated multiple molecules in the roles as effectors of staphylococcal biofilm proliferation/structuring and detachment/dispersal, to date, only phenol soluble modulins (PSMs) have been consistently demonstrated to serve in this role under both in vitro and in vivo settings. PSMs are regulated directly through a density-dependent manner by the accessory gene regulator (Agr) system. They disrupt the non-covalent forces holding the biofilm extracellular matrix together, which is necessary for the formation of channels, a process essential for the delivery of nutrients to deeper biofilm layers, and for dispersal/dissemination of clusters of biofilm to distal organs in acute infection. Given their relevance in both acute and chronic biofilm-associated infections, the Agr system and the psm genes hold promise as potential therapeutic targets.
Keywords: Staphylococcus aureus; Staphylococcus epidermidis; biofilm; medical devices; phenol-soluble modulins.
Figures


References
-
- Cheung G. Y., Kretschmer D., Queck S. Y., Joo H. S., Wang R., Duong A. C., et al. . (2014b). Insight into structure-function relationship in phenol-soluble modulins using an alanine screen of the phenol-soluble modulin (PSM) alpha3 peptide. FASEB J. 28, 153–161. 10.1096/fj.13-232041 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

