Concepts, challenges, and successes in modeling thermodynamics of metabolism
- PMID: 25505786
- PMCID: PMC4244978
- DOI: 10.3389/fbioe.2014.00053
Concepts, challenges, and successes in modeling thermodynamics of metabolism
Abstract
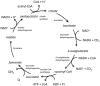
The modeling of the chemical reactions involved in metabolism is a daunting task. Ideally, the modeling of metabolism would use kinetic simulations, but these simulations require knowledge of the thousands of rate constants involved in the reactions. The measurement of rate constants is very labor intensive, and hence rate constants for most enzymatic reactions are not available. Consequently, constraint-based flux modeling has been the method of choice because it does not require the use of the rate constants of the law of mass action. However, this convenience also limits the predictive power of constraint-based approaches in that the law of mass action is used only as a constraint, making it difficult to predict metabolite levels or energy requirements of pathways. An alternative to both of these approaches is to model metabolism using simulations of states rather than simulations of reactions, in which the state is defined as the set of all metabolite counts or concentrations. While kinetic simulations model reactions based on the likelihood of the reaction derived from the law of mass action, states are modeled based on likelihood ratios of mass action. Both approaches provide information on the energy requirements of metabolic reactions and pathways. However, modeling states rather than reactions has the advantage that the parameters needed to model states (chemical potentials) are much easier to determine than the parameters needed to model reactions (rate constants). Herein, we discuss recent results, assumptions, and issues in using simulations of state to model metabolism.
Keywords: adaptation; biological; fluctuation theory; metabolism; molecular motors; simulations; statistical thermodynamics; tricarboxylic acid cycle.
Figures

Similar articles
-
Simulating metabolism with statistical thermodynamics.PLoS One. 2014 Aug 4;9(8):e103582. doi: 10.1371/journal.pone.0103582. eCollection 2014. PLoS One. 2014. PMID: 25089525 Free PMC article.
-
Evaluation of rate law approximations in bottom-up kinetic models of metabolism.BMC Syst Biol. 2016 Jun 6;10(1):40. doi: 10.1186/s12918-016-0283-2. BMC Syst Biol. 2016. PMID: 27266508 Free PMC article.
-
Non-steady state mass action dynamics without rate constants: dynamics of coupled reactions using chemical potentials.Phys Biol. 2017 Aug 16;14(5):055003. doi: 10.1088/1478-3975/aa7d80. Phys Biol. 2017. PMID: 28675379
-
Parameters for pyrethroid insecticide QSAR and PBPK/PD models for human risk assessment.Rev Environ Contam Toxicol. 2012;219:1-114. doi: 10.1007/978-1-4614-3281-4_1. Rev Environ Contam Toxicol. 2012. PMID: 22610175 Review.
-
Shrinking the metabolic solution space using experimental datasets.PLoS Comput Biol. 2012;8(8):e1002662. doi: 10.1371/journal.pcbi.1002662. Epub 2012 Aug 30. PLoS Comput Biol. 2012. PMID: 22956899 Free PMC article. Review.
Cited by
-
Editorial: Current Challenges in Modeling Cellular Metabolism.Front Bioeng Biotechnol. 2015 Nov 26;3:193. doi: 10.3389/fbioe.2015.00193. eCollection 2015. Front Bioeng Biotechnol. 2015. PMID: 26636080 Free PMC article. No abstract available.
-
The Energy Homeostasis Principle: Neuronal Energy Regulation Drives Local Network Dynamics Generating Behavior.Front Comput Neurosci. 2019 Jul 23;13:49. doi: 10.3389/fncom.2019.00049. eCollection 2019. Front Comput Neurosci. 2019. PMID: 31396067 Free PMC article.
-
Energy-harnessing problem solving of primordial life: Modeling the emergence of catalytic host-nested parasite life cycles.PLoS One. 2023 Mar 27;18(3):e0281661. doi: 10.1371/journal.pone.0281661. eCollection 2023. PLoS One. 2023. PMID: 36972235 Free PMC article.
-
Overcoming substrate limitations for improved production of ethylene in E. coli.Biotechnol Biofuels. 2016 Jan 4;9:3. doi: 10.1186/s13068-015-0413-x. eCollection 2016. Biotechnol Biofuels. 2016. PMID: 26734073 Free PMC article.
References
-
- Barato A. C., Hartich D., Seifert U. (2014). Efficiency of cellular information processing. New J. Phys. 16, 103024.10.1088/1367-2630/16/10/103024 - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources

