Developments in the tools and methodologies of synthetic biology
- PMID: 25505788
- PMCID: PMC4244866
- DOI: 10.3389/fbioe.2014.00060
Developments in the tools and methodologies of synthetic biology
Abstract
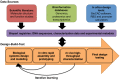
Synthetic biology is principally concerned with the rational design and engineering of biologically based parts, devices, or systems. However, biological systems are generally complex and unpredictable, and are therefore, intrinsically difficult to engineer. In order to address these fundamental challenges, synthetic biology is aiming to unify a "body of knowledge" from several foundational scientific fields, within the context of a set of engineering principles. This shift in perspective is enabling synthetic biologists to address complexity, such that robust biological systems can be designed, assembled, and tested as part of a biological design cycle. The design cycle takes a forward-design approach in which a biological system is specified, modeled, analyzed, assembled, and its functionality tested. At each stage of the design cycle, an expanding repertoire of tools is being developed. In this review, we highlight several of these tools in terms of their applications and benefits to the synthetic biology community.
Keywords: design cycle; engineering biology; standardization; synthetic biology; tools.
Figures
Similar articles
-
Design Methodologies and the Limits of the Engineering-Dominated Conception of Synthetic Biology.Acta Biotheor. 2019 Mar;67(1):1-18. doi: 10.1007/s10441-018-9338-7. Epub 2018 Aug 18. Acta Biotheor. 2019. PMID: 30121875
-
Synthetic biology: an emerging engineering discipline.Annu Rev Biomed Eng. 2012;14:155-78. doi: 10.1146/annurev-bioeng-071811-150118. Epub 2012 May 7. Annu Rev Biomed Eng. 2012. PMID: 22577777 Review.
-
Tools and Principles for Microbial Gene Circuit Engineering.J Mol Biol. 2016 Feb 27;428(5 Pt B):862-88. doi: 10.1016/j.jmb.2015.10.004. Epub 2015 Oct 20. J Mol Biol. 2016. PMID: 26463592 Review.
-
Recent Advances and Current Challenges in Synthetic Biology of the Plastid Genetic System and Metabolism.Plant Physiol. 2019 Mar;179(3):794-802. doi: 10.1104/pp.18.00767. Epub 2018 Sep 4. Plant Physiol. 2019. PMID: 30181342 Free PMC article. Review.
-
Plant synthetic biology.Trends Plant Sci. 2015 May;20(5):309-317. doi: 10.1016/j.tplants.2015.02.004. Epub 2015 Mar 27. Trends Plant Sci. 2015. PMID: 25825364 Review.
Cited by
-
A forward-design approach to increase the production of poly-3-hydroxybutyrate in genetically engineered Escherichia coli.PLoS One. 2015 Feb 20;10(2):e0117202. doi: 10.1371/journal.pone.0117202. eCollection 2015. PLoS One. 2015. PMID: 25699671 Free PMC article.
-
Analytics for Metabolic Engineering.Front Bioeng Biotechnol. 2015 Sep 7;3:135. doi: 10.3389/fbioe.2015.00135. eCollection 2015. Front Bioeng Biotechnol. 2015. PMID: 26442249 Free PMC article. Review.
-
Construction of a Calibration Curve for Lycopene on a Liquid-Handling Platform─Wider Lessons for the Development of Automated Dilution Protocols.ACS Synth Biol. 2024 Aug 16;13(8):2357-2375. doi: 10.1021/acssynbio.4c00031. Epub 2024 Aug 3. ACS Synth Biol. 2024. PMID: 39096303 Free PMC article.
-
Standardization of Synthetic Biology Tools and Assembly Methods for Saccharomyces cerevisiae and Emerging Yeast Species.ACS Synth Biol. 2022 Aug 19;11(8):2527-2547. doi: 10.1021/acssynbio.1c00442. Epub 2022 Aug 8. ACS Synth Biol. 2022. PMID: 35939789 Free PMC article. Review.
-
Molecular circuits for dynamic noise filtering.Proc Natl Acad Sci U S A. 2016 Apr 26;113(17):4729-34. doi: 10.1073/pnas.1517109113. Epub 2016 Apr 12. Proc Natl Acad Sci U S A. 2016. PMID: 27078094 Free PMC article.
References
-
- Anderson J., Strelkowa N., Stan G. B., Douglas T., Savulescu J., Barahona M., et al. (2012). Engineering and ethical perspectives in synthetic biology. Rigorous, robust and predictable designs, public engagement and a modern ethical framework are vital to the continued success of synthetic biology. EMBO Rep. 13, 584–59010.1038/embor.2012.81 - DOI - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

