Comparative proteomic analysis of indica and japonica rice varieties
- PMID: 25505840
- PMCID: PMC4261965
- DOI: 10.1590/S1415-47572014005000015
Comparative proteomic analysis of indica and japonica rice varieties
Abstract
Indica and japonica are two main subspecies of Asian cultivated rice (Oryza sativa L.) that differ clearly in morphological and agronomic traits, in physiological and biochemical characteristics and in their genomic structure. However, the proteins and genes responsible for these differences remain poorly characterized. In this study, proteomic tools, including two-dimensional electrophoresis and mass spectrometry, were used to globally identify proteins that differed between two sequenced rice varieties (93-11 and Nipponbare). In all, 47 proteins that differed significantly between 93-11 and Nipponbare were identified using mass spectrometry and database searches. Interestingly, seven proteins were expressed only in Nipponbare and one protein was expressed specifically in 93-11; these differences were confirmed by quantitative real-time PCR and proteomic analysis of other indica and japonica rice varieties. This is the first report to successfully demonstrate differences in the protein composition of indica and japonica rice varieties and to identify candidate proteins and genes for future investigation of their roles in the differentiation of indica and japonica rice.
Keywords: indica and japonica rice; molecular marker; proteomics; quantitative real-time PCR; unique proteins.
Figures
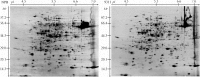


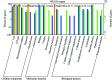
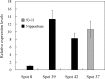

References
-
- Agrawal GK, Rakwal R. Rice proteomics: A move toward expanded proteome coverage to comparative and functional proteomics uncovers the mysteries of rice and plant biology. Proteomics. 2011;11:1630–1649. - PubMed
-
- Agrawal GK, Jwa NS, Iwahashi Y, Yonekura M, Iwahashi H, Rakwal R. Rejuvenating rice proteomics: Facts, challenges, and visions. Proteomics. 2006;6:5549–5576. - PubMed
-
- Bevan M, Bancroft I, Bent E, Love K, Goodman H, Dean C, Bergkamp R, Dirkse W, Van Staveren M, Stiekema W, et al. Analysis of 1.9 Mb of contiguous sequence from chromosome 4 of Arabidopsis thaliana. Nature. 1998;391:485–488. - PubMed
-
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. - PubMed
-
- Chi F, Yang P, Han F, Jing Y, Shen S. Proteomic analysis of rice seedlings infected by Sinorhizobium meliloti 1021. Proteomics. 2010;10:1861–1874. - PubMed
Internet Resources
-
- Mascot program, http://www.matrixscience.com November 30, 2012.
-
- InterProscan program, http://www.ebi.ac.uk/Tools/InterProScan January 9, 2013.
-
- WEGO software, http://wego.genomics.org.cn January 10, 2013.
LinkOut - more resources
Full Text Sources
Other Literature Sources

