Transcriptome Analysis of Early Responsive Genes in Rice during Magnaporthe oryzae Infection
- PMID: 25506299
- PMCID: PMC4262287
- DOI: 10.5423/PPJ.OA.06.2014.0055
Transcriptome Analysis of Early Responsive Genes in Rice during Magnaporthe oryzae Infection
Abstract
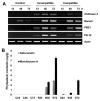
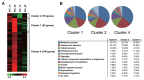
Rice blast disease caused by Magnaporthe oryzae is one of the most serious diseases of cultivated rice (Oryza sativa L.) in most rice-growing regions of the world. In order to investigate early response genes in rice, we utilized the transcriptome analysis approach using a 300 K tilling microarray to rice leaves infected with compatible and incompatible M. oryzae strains. Prior to the microarray experiment, total RNA was validated by measuring the differential expression of rice defense-related marker genes (chitinase 2, barwin, PBZ1, and PR-10) by RT-PCR, and phytoalexins (sakuranetin and momilactone A) with HPLC. Microarray analysis revealed that 231 genes were up-regulated (>2 fold change, p < 0.05) in the incompatible interaction compared to the compatible one. Highly expressed genes were functionally characterized into metabolic processes and oxidation-reduction categories. The oxidative stress response was induced in both early and later infection stages. Biotic stress overview from MapMan analysis revealed that the phytohormone ethylene as well as signaling molecules jasmonic acid and salicylic acid is important for defense gene regulation. WRKY and Myb transcription factors were also involved in signal transduction processes. Additionally, receptor-like kinases were more likely associated with the defense response, and their expression patterns were validated by RT-PCR. Our results suggest that candidate genes, including receptor-like protein kinases, may play a key role in disease resistance against M. oryzae attack.
Keywords: Magnaporthe oryzae; MapMan analysis; defense response; rice; transcriptomics.
Figures



References
-
- Acharya BR, Raina S, Maqbool SB, Jagadeeswaran G, Mosher SL, Appel HM, Schultz JC, Klessig DF, Raina R. Overexpression of CRK13, an Arabidopsis cysteine-rich receptor-like kinase, results in enhanced resistance to Pseudomonas syringae. Plant J. 2007;50:488–499. - PubMed
-
- Arlorio M, Ludwig A, Boller T, Bonfante P. Inhibition of fungal growth by plant chitinases and β-1,3-glucanases. Protoplasma. 1992;171:34–43.
-
- Bagnaresi P, Biselli C, Orru L, Urso S, Crispino L, Abbruscato P, Piffanelli P, Lupotto E, Cattivelli L, Vale G. Comparative transcriptome profiling of the early response to Magnaporthe oryzae in durable resistant vs susceptible rice (Oryza sativa L.) genotypes. PLoS One. 2012;7:e51609. - PMC - PubMed
-
- Bennett RN, Wallsgrove RM. Secondary metabolites in plant defence mechanisms. New Phytol. 1994;127:617–633. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

