Gene prediction and annotation in Penstemon (Plantaginaceae): A workflow for marker development from extremely low-coverage genome sequencing
- PMID: 25506519
- PMCID: PMC4259454
- DOI: 10.3732/apps.1400044
Gene prediction and annotation in Penstemon (Plantaginaceae): A workflow for marker development from extremely low-coverage genome sequencing
Abstract
Premise of the study: Penstemon (Plantaginaceae) is a large and diverse genus endemic to North America. However, determining the phylogenetic relationships among its 280 species has been difficult due to its recent evolutionary radiation. The development of a large, multilocus data set can help to resolve this challenge. •
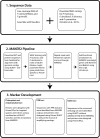
Methods: Using both previously sequenced genomic libraries and our own low-coverage whole-genome shotgun sequencing libraries, we used the MAKER2 Annotation Pipeline to identify gene regions for the development of sequencing loci from six extremely low-coverage Penstemon genomes (∼0.005×-0.007×). We also compared this approach to BLAST searches, and conducted analyses to characterize sequence divergence across the species sequenced. •
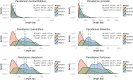
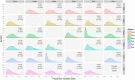
Results: Annotations and gene predictions were successfully added to more than 10,000 contigs for potential use in downstream primer design. Primers were then designed for chloroplast, mitochondrial, and nuclear loci from these annotated sequences. MAKER2 identified longer gene regions in all six Penstemon genomes when compared with BLASTN and BLASTX searches. The average level of sequence divergence among the six species was 7.14%. •
Discussion: Combining bioinformatics tools into a workflow that produces annotations can be useful for creating potential phylogenetic markers from thousands of sequences even when genome coverage is extremely low and reference data are only available from distant relatives. Furthermore, the output from MAKER2 contains information about important gene features, such as exon boundaries, and can be easily integrated with visualization tools to facilitate the process of marker development.
Keywords: 454 pyrosequencing; BLAST; MAKER2; Penstemon; bioinformatics.
Figures
Similar articles
-
Developing molecular tools and insights into the Penstemon genome using genomic reduction and next-generation sequencing.BMC Genet. 2013 Aug 8;14:66. doi: 10.1186/1471-2156-14-66. BMC Genet. 2013. PMID: 23924218 Free PMC article.
-
Multiplexed shotgun genotyping resolves species relationships within the North American genus Penstemon.Am J Bot. 2016 May;103(5):912-22. doi: 10.3732/ajb.1500519. Epub 2016 May 10. Am J Bot. 2016. PMID: 27208359 Free PMC article.
-
Phylogeny, taxonomic affinities, and biogeography of Penstemon (Plantaginaceae) based on ITS and cpDNA sequence data.Am J Bot. 2006 Nov;93(11):1699-713. doi: 10.3732/ajb.93.11.1699. Am J Bot. 2006. PMID: 21642115
-
MAKER2: an annotation pipeline and genome-database management tool for second-generation genome projects.BMC Bioinformatics. 2011 Dec 22;12:491. doi: 10.1186/1471-2105-12-491. BMC Bioinformatics. 2011. PMID: 22192575 Free PMC article.
-
Whole-genome sequence and annotation of Penstemon davidsonii.G3 (Bethesda). 2024 Mar 6;14(3):jkad296. doi: 10.1093/g3journal/jkad296. G3 (Bethesda). 2024. PMID: 38155402 Free PMC article.
Cited by
-
The unexpected depths of genome-skimming data: A case study examining Goodeniaceae floral symmetry genes.Appl Plant Sci. 2017 Oct 20;5(10):apps.1700042. doi: 10.3732/apps.1700042. eCollection 2017 Oct. Appl Plant Sci. 2017. PMID: 29109919 Free PMC article.
-
The Complete Plastome Sequence Of Penstemon fruticosus (Pursh) Greene (Plantaginaceae).Mitochondrial DNA B Resour. 2017 Nov 6;2(2):768-769. doi: 10.1080/23802359.2017.1398620. Mitochondrial DNA B Resour. 2017. PMID: 33473975 Free PMC article.
-
Primers for Castilleja and their utility across Orobanchaceae: II. Single-copy nuclear loci.Appl Plant Sci. 2017 Sep 30;5(9):apps.1700038. doi: 10.3732/apps.1700038. eCollection 2017 Sep. Appl Plant Sci. 2017. PMID: 28989822 Free PMC article.
-
Congruent Deep Relationships in the Grape Family (Vitaceae) Based on Sequences of Chloroplast Genomes and Mitochondrial Genes via Genome Skimming.PLoS One. 2015 Dec 14;10(12):e0144701. doi: 10.1371/journal.pone.0144701. eCollection 2015. PLoS One. 2015. PMID: 26656830 Free PMC article.
-
Low-coverage, whole-genome sequencing of Artocarpus camansi (Moraceae) for phylogenetic marker development and gene discovery.Appl Plant Sci. 2016 Jul 13;4(7):apps.1600017. doi: 10.3732/apps.1600017. eCollection 2016 Jul. Appl Plant Sci. 2016. PMID: 27437173 Free PMC article.
References
-
- Altschul S. F., Gish W., Miller W., Meyer E. W., Lipman D. J. 1990. Basic local alignment search tool. Journal of Molecular Biology 215: 403–410. - PubMed
-
- Blischak P. D., Wenzel A. J., Wolfe A. D. 2014. Data from: Gene prediction and annotation in Penstemon (Plantaginaceae): A workflow for marker development from extremely low-coverage genome sequencing. Dryad Digital Repository. http://doi.org/10.5061/dryad.f6s22. - DOI - PMC - PubMed
-
- Broderick S. R., Stevens M. R., Geary B., Love S. L., Jellen E. N., Dockter R. B., Daley S. L., Lindgren D. T. 2011. A survey of Penstemon’s genome size. Genome 54: 160–173. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

