L1CAM is expressed in triple-negative breast cancers and is inversely correlated with androgen receptor
- PMID: 25510351
- PMCID: PMC4301892
- DOI: 10.1186/1471-2407-14-958
L1CAM is expressed in triple-negative breast cancers and is inversely correlated with androgen receptor
Abstract
Background: Breast cancer is a heterogeneous disease displaying distinct molecular features and clinical outcome. The molecular profile of triple-negative breast cancers (TNBCs) overlaps with that of basal-like breast cancers that in turn show similarities with high-grade serous ovarian and endometrial carcinoma. L1CAM is an established biomarker for the latter cancers and we showed before that approximately 18% of primary breast cancers are positive for L1CAM and have a bad prognosis. Here we analysed the expression of L1CAM breast cancer subtypes.
Methods: We analyzed mRNA and protein expression data from different breast cancer cohorts for L1CAM, estrogen receptor, progesterone receptor, Her-2 and Androgen receptor (AR) and correlated the data. We performed Western blot analysis on tumor cell lysates and carried out chromatin-immuno-precipitation (CHIP) after AR overexpression.
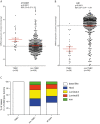
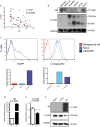
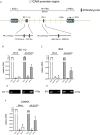
Results: We find that L1CAM is expressed preferentially though not exclusively in TNBCs. Using the human cancer genome atlas database and two independent breast cancer cohorts we find that L1CAM is inversely correlated with androgen receptor (AR) expression. We found that L1CAM(high)AR(low) primary breast tumors have the worst clinical outcome. Overexpression of AR in MDA-MB436 breast cancer cells decreased L1CAM expression at the protein and mRNA level and CHIP-analysis revealed binding of AR to the L1CAM promoter region.
Conclusions: These results suggest that L1CAM in breast cancer is under AR control. The data also strongly advocate the use of L1CAM assessment in breast cancer diagnosis. We suggest that L1CAM expression could be causally related to the bad prognosis of TNBCs.
Figures



References
-
- Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, Fluge O, Pergamenschikov A, Williams C, Zhu SX, Lonning PE, Borresen-Dale AL, Brown PO, Botstein D. Molecular portraits of human breast tumours. Nature. 2000;406(6797):747–752. doi: 10.1038/35021093. - DOI - PubMed
-
- Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey SS, Thorsen T, Quist H, Matese JC, Brown PO, Botstein D, Lonning PE, Borresen-Dale AL. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98(19):10869–10874. doi: 10.1073/pnas.191367098. - DOI - PMC - PubMed
Pre-publication history
-
- The pre-publication history for this paper can be accessed here: http://www.biomedcentral.com/1471-2407/14/958/prepub
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

