Mycobacterial RNA polymerase forms unstable open promoter complexes that are stabilized by CarD
- PMID: 25510492
- PMCID: PMC4288152
- DOI: 10.1093/nar/gku1231
Mycobacterial RNA polymerase forms unstable open promoter complexes that are stabilized by CarD
Abstract
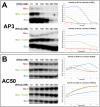
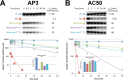
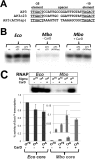
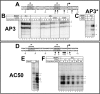
Escherichia coli has served as the archetypal organism on which the overwhelming majority of biochemical characterizations of bacterial RNA polymerase (RNAP) have been focused; the properties of E. coli RNAP have been accepted as generally representative for all bacterial RNAPs. Here, we directly compare the initiation properties of a mycobacterial transcription system with E. coli RNAP on two different promoters. The detailed characterizations include abortive transcription assays, RNAP/promoter complex stability assays and DNAse I and KMnO4 footprinting. Based on footprinting, we find that promoter complexes formed by E. coli and mycobacterial RNAPs use very similar protein/DNA interactions and generate the same transcription bubbles. However, we find that the open promoter complexes formed by E. coli RNAP on the two promoters tested are highly stable and essentially irreversible (with lifetimes much greater than 1 h), while the open promoter complexes on the same two promoters formed by mycobacterial RNAP are very unstable (lifetimes of about 2 min or less) and readily reversible. We show here that CarD, an essential mycobacterial transcription activator that is not found in E. coli, stabilizes the mycobacterial RNAP/open promoter complexes considerably by preventing transcription bubble collapse.
© The Author(s) 2014. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures



References
-
- Floss H.G., Yu T.-W. Rifamycin-mode of action, resistance, and biosynthesis. Chem. Rev. 2005;105:621–632. - PubMed
-
- Burgess R.R. Separation and characterization of the subunits of ribonucleic acid polymerase. J. Biol. Chem. 1969;244:6168–6176. - PubMed
-
- Burgess R.R., Travers A.A., Dunn J.J., Bautz E.K. Factor stimulating transcription by RNA polymerase. Nature. 1969;221:43–46. - PubMed
-
- Burgess R.R., Travers A.A. Escherichia coli RNA polymerase: purification, subunit structure, and factor requirements. Fed. Proc. 1970;29:1164–1169. - PubMed
-
- Zhang G., Campbell E.A., Minakhin L., Richter C., Severinov K., Darst S.A. Crystal structure of Thermus aquaticus core RNA polymerase at 3.3 A resolution. Cell. 1999;98:811–824. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

