Comparisons of computational methods for differential alternative splicing detection using RNA-seq in plant systems
- PMID: 25511303
- PMCID: PMC4271460
- DOI: 10.1186/s12859-014-0364-4
Comparisons of computational methods for differential alternative splicing detection using RNA-seq in plant systems
Abstract
Background: Alternative Splicing (AS) as a post-transcription regulation mechanism is an important application of RNA-seq studies in eukaryotes. A number of software and computational methods have been developed for detecting AS. Most of the methods, however, are designed and tested on animal data, such as human and mouse. Plants genes differ from those of animals in many ways, e.g., the average intron size and preferred AS types. These differences may require different computational approaches and raise questions about their effectiveness on plant data. The goal of this paper is to benchmark existing computational differential splicing (or transcription) detection methods so that biologists can choose the most suitable tools to accomplish their goals.
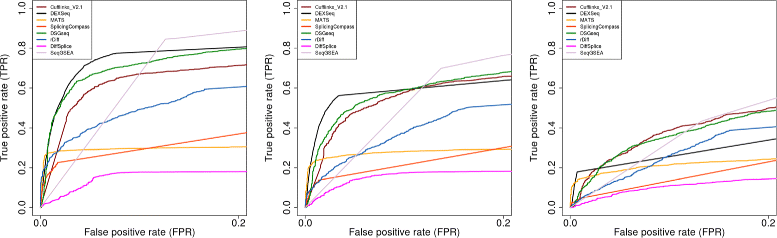
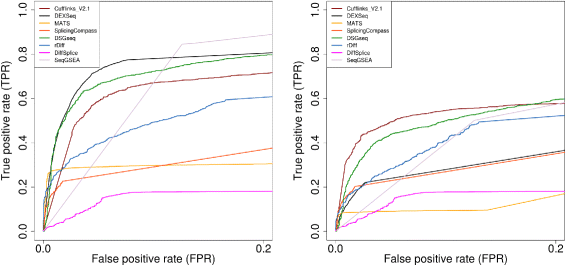
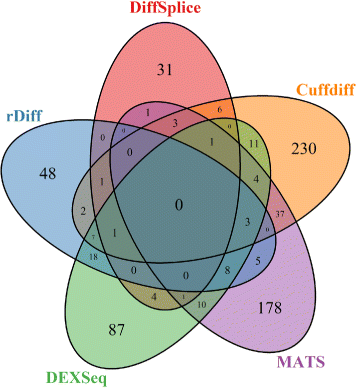
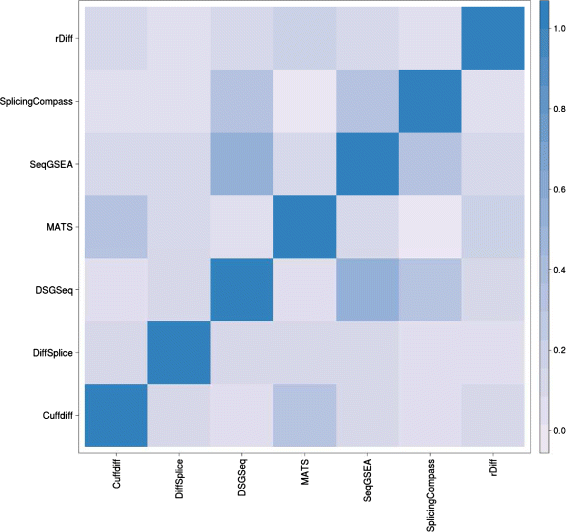
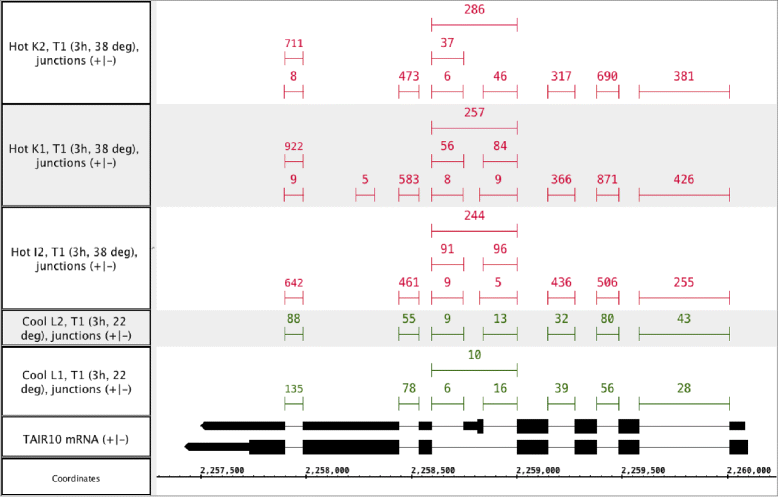
Results: This study compares the eight popular public available software packages for differential splicing analysis using both simulated and real Arabidopsis thaliana RNA-seq data. All software are freely available. The study examines the effect of varying AS ratio, read depth, dispersion pattern, AS types, sample sizes and the influence of annotation. Using a real data, the study looks at the consistences between the packages and verifies a subset of the detected AS events using PCR studies.
Conclusions: No single method performs the best in all situations. The accuracy of annotation has a major impact on which method should be chosen for AS analysis. DEXSeq performs well in the simulated data when the AS signal is relative strong and annotation is accurate. Cufflinks achieve a better tradeoff between precision and recall and turns out to be the best one when incomplete annotation is provided. Some methods perform inconsistently for different AS types. Complex AS events that combine several simple AS events impose problems for most methods, especially for MATS. MATS stands out in the analysis of real RNA-seq data when all the AS events being evaluated are simple AS events.
Figures
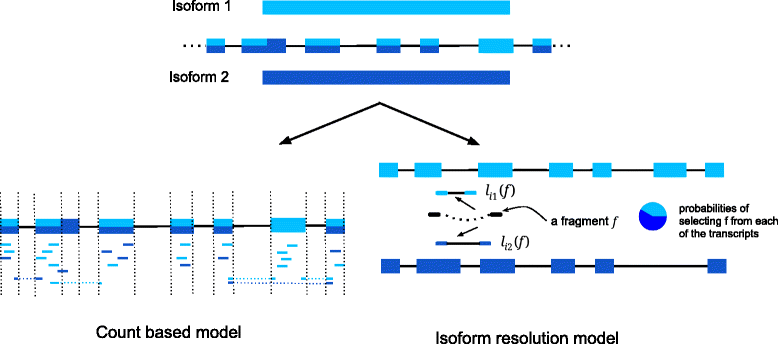

References
-
- Graveley BR, Brooks AN, Carlson JW, Duff MO, Landolin JM, Yang L, Artieri CG, van Baren MJ, Boley N, Booth BW, Brown JB, Cherbas L, Davis CA, Dobin A, Li R, Lin W, Malone JH, Mattiuzzo NR, Miller D, Sturgill D, Tuch BB, Zaleski C, Zhang D, Blanchette M, Dudoit S, Eads B, Green RE, Hammonds A, Jiang L, Kapranov P, et al. The developmental transcriptome of Drosophila melanogaster. Nature. 2011;471(7339):473–479. doi: 10.1038/nature09715. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

