Spatial genome organization: contrasting views from chromosome conformation capture and fluorescence in situ hybridization
- PMID: 25512564
- PMCID: PMC4265680
- DOI: 10.1101/gad.251694.114
Spatial genome organization: contrasting views from chromosome conformation capture and fluorescence in situ hybridization
Abstract
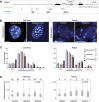
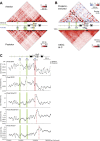
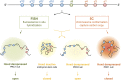
Although important for gene regulation, most studies of genome organization use either fluorescence in situ hybridization (FISH) or chromosome conformation capture (3C) methods. FISH directly visualizes the spatial relationship of sequences but is usually applied to a few loci at a time. The frequency at which sequences are ligated together by formaldehyde cross-linking can be measured genome-wide by 3C methods, with higher frequencies thought to reflect shorter distances. FISH and 3C should therefore give the same views of genome organization, but this has not been tested extensively. We investigated the murine HoxD locus with 3C carbon copy (5C) and FISH in different developmental and activity states and in the presence or absence of epigenetic regulators. We identified situations in which the two data sets are concordant but found other conditions under which chromatin topographies extrapolated from 5C or FISH data are not compatible. We suggest that products captured by 3C do not always reflect spatial proximity, with ligation occurring between sequences located hundreds of nanometers apart, influenced by nuclear environment and chromatin composition. We conclude that results obtained at high resolution with either 3C methods or FISH alone must be interpreted with caution and that views about genome organization should be validated by independent methods.
Keywords: 3C; FISH; Hox genes; nuclear organization; polycomb.
© 2014 Williamson et al.; Published by Cold Spring Harbor Laboratory Press.
Figures



References
-
- Albiez H, Cremer M, Tiberi C, Vecchio L, Schermelleh L, Dittrich S, Küpper K, Joffe B, Thormeyer T, von Hase J, et al. . 2006. Chromatin domains and the interchromatin compartment form structurally defined and functionally interacting nuclear networks. Chrom Res 14: 707–733. - PubMed
-
- Amano T, Sagai T, Tanabe H, Mizushina Y, Nakazawa H, Shiroishi T. 2009. Chromosomal dynamics at the Shh locus: limb bud-specific differential regulation of competence and active transcription. Dev Cell 16: 47–57. - PubMed
-
- Andrey G, Montavon T, Mascrez B, Gonzalez F, Noordermeer D, Leleu M, Trono D, Spitz F, Duboule D. 2013. A switch between topological domains underlies HoxD genes collinearity in mouse limbs. Science 340: 1234167. - PubMed
-
- Azuara V, Perry P, Sauer S, Spivakov M, Jørgensen HF, John RM, Gouti M, Casanova M, Warnes G, Merkenschlager M, et al. . 2006. Chromatin signatures of pluripotent cell lines. Nat Cell Biol 8: 532–538. - PubMed
-
- Baù D, Marti-Renom MA. 2011. Structure determination of genomic domains by satisfaction of spatial restraints. Chrom Res 19: 25–35. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
